Carboxylesterase
Carboxylesterases hydrolyse carboxylic ester bonds with relatively broad substrate specificity and are useful for stereospecific synthesis and hydrolysis of esters.
Reference Protein and Structure
- Sequence
-
Q0ZPV7
 (3.1.1.1)
(3.1.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Actinidia eriantha (Kiwi)

- PDB
-
2o7r
- Plant carboxylesterase AeCXE1 from Actinidia eriantha with acyl adduct
(1.4 Å)



- Catalytic CATH Domains
-
3.40.50.1820
 (see all for 2o7r)
(see all for 2o7r)
Enzyme Reaction (EC:3.1.1.1)
Enzyme Mechanism
Introduction
Carboxylesterases use a Ser-His-Asp catalytic triad. Serine, deprotonated by histidine which is stabilised in its charged form by aspartate, carries out a nucleophilic attack on the carbonyl of the substrate. The resulting tetrahedral transition state is stabilised by an oxyanion hole. When it collapses, histidine protonates the leaving group and the result is a covalent enzyme-substrate intermediate. A water molecule, deprotonated by histidine, attacks the covalent intermediate to give another tetrahedral intermediate, which collapses to give the product and the enzyme in its native state.
Catalytic Residues Roles
| UniProt | PDB* (2o7r) | ||
| Gly92 (main-N), Gly93 (main-N), Ala170 (main-N) | Gly92A (main-N), Gly93A (main-N), Ala170A (main-N) | Oxyanion hole formed by the backbone amine groups of Gly92, Gly93 and Ala170. The ester substrate is stabilised in the active site by three hydrogen bonds from the NH groups of the oxyanion hole. These bonds align the substrate with Ser169 which is essential for initiating the catalytic process. | hydrogen bond donor, electrostatic stabiliser |
| Ser169, His306, Asp276 | Ser169A, His306A, Asp276A | Ser169, Asp276 and His306 form a conserved catalytic triad of residues. Ser169 performs a nucleophilic attack on the substrate and forms the acyl-enzyme complex. His306 acts as a general acid/base, facilitating the formation and cleavage of the two tetrahedral intermediates. Asp276 is hydrogen-bonded to His306, Gln282 and Met278. | hydrogen bond acceptor, hydrogen bond donor, nucleophile, nucleofuge, covalent catalysis |
| Gln282, Met278 (main-N) | Gln282A, Met278A (main-N) | Gln282 and Met278 are hydrogen-bonded to Asp276. | hydrogen bond donor |
Chemical Components
bimolecular nucleophilic addition, proton transfer, overall reactant used, intermediate formation, enzyme-substrate complex formation, intermediate collapse, unimolecular elimination by the conjugate base, overall product formed, heterolysis, enzyme-substrate complex cleavage, native state of enzyme regeneratedReferences
- Aranda J et al. (2014), Biochemistry, 53, 5820-5829. The catalytic mechanism of carboxylesterases: a computational study. DOI:10.1021/bi500934j. PMID:25101647.
- Yan M et al. (2021), Comput Theor Chem, 1199, 113198-. Catalytic hydrolysis mechanism of aminocarboxylester substrate by human carboxylesterase 1: A theoretical study on methylphenidate hydrolysis. DOI:10.1016/j.comptc.2021.113198.
- Zhan H et al. (2020), Environ Res, 182, 109138-. New insights into the microbial degradation and catalytic mechanism of synthetic pyrethroids. DOI:10.1016/j.envres.2020.109138. PMID:32069744.
- Yan M et al. (2019), Molecules, 24,Catalytic Hydrolysis Mechanism of Cocaine by Human Carboxylesterase 1: An Orthoester Intermediate Slows Down the Reaction. DOI:10.3390/molecules24224057. PMID:31717501.
- Kim KK et al. (1997), Structure, 5, 1571-1584. Crystal structure of carboxylesterase from Pseudomonas fluorescens, an α/β hydrolase with broad substrate specificity. DOI:10.1016/s0969-2126(97)00306-7. PMID:9438866.

Step 1. The carbonyl group forms three hydrogen bonds to the backbone nitrogen atoms of Gly92 and Gly93 of the oxyanion hole region and the backbone nitrogen of two of them with the backbone nitrogen atoms of Ala170. His306 facilitates the nucleophilic attack of Ser169 to the substrate. The first tetrahedral intermediate forms.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly92A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Gly93A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Ala170A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Ser169A | hydrogen bond donor |
| His306A | hydrogen bond acceptor, hydrogen bond donor |
| Asp276A | hydrogen bond acceptor |
| Gln282A | hydrogen bond donor |
| Met278A (main-N) | hydrogen bond donor |
| His306A | increase nucleophilicity, electrostatic stabiliser |
| Asp276A | electrostatic stabiliser |
| Ser169A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, intermediate formation, enzyme-substrate complex formation
Step 2. Collapse of tetrahedral intermediate to an acyl-enzyme complex and release of alcohol product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly92A (main-N) | hydrogen bond donor |
| Gly93A (main-N) | hydrogen bond donor |
| Ala170A (main-N) | hydrogen bond donor |
| Ser169A | hydrogen bond donor |
| His306A | hydrogen bond donor |
| Gln282A | hydrogen bond donor |
| Met278A (main-N) | hydrogen bond donor |
| Asp276A | hydrogen bond acceptor |
| Ser169A | covalent catalysis |
| His306A | promote heterolysis |
Chemical Components
proton transfer, intermediate collapse, ingold: unimolecular elimination by the conjugate base, overall product formed, heterolysis
Step 3. A water molecule enters the active site. Nucleophilic attack of a water molecule on the acyl-enzyme complex facilitated by His306. A second tetrahedral intermediate forms.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His306A | hydrogen bond acceptor, hydrogen bond donor |
| Ser169A | hydrogen bond acceptor |
| Asp276A | hydrogen bond acceptor |
| Gly92A (main-N) | hydrogen bond donor |
| Gly93A (main-N) | hydrogen bond donor |
| Ala170A (main-N) | hydrogen bond donor |
| Gln282A | hydrogen bond donor |
| Met278A (main-N) | hydrogen bond donor |
| His306A | increase nucleophilicity |
| Ser169A | covalent catalysis |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation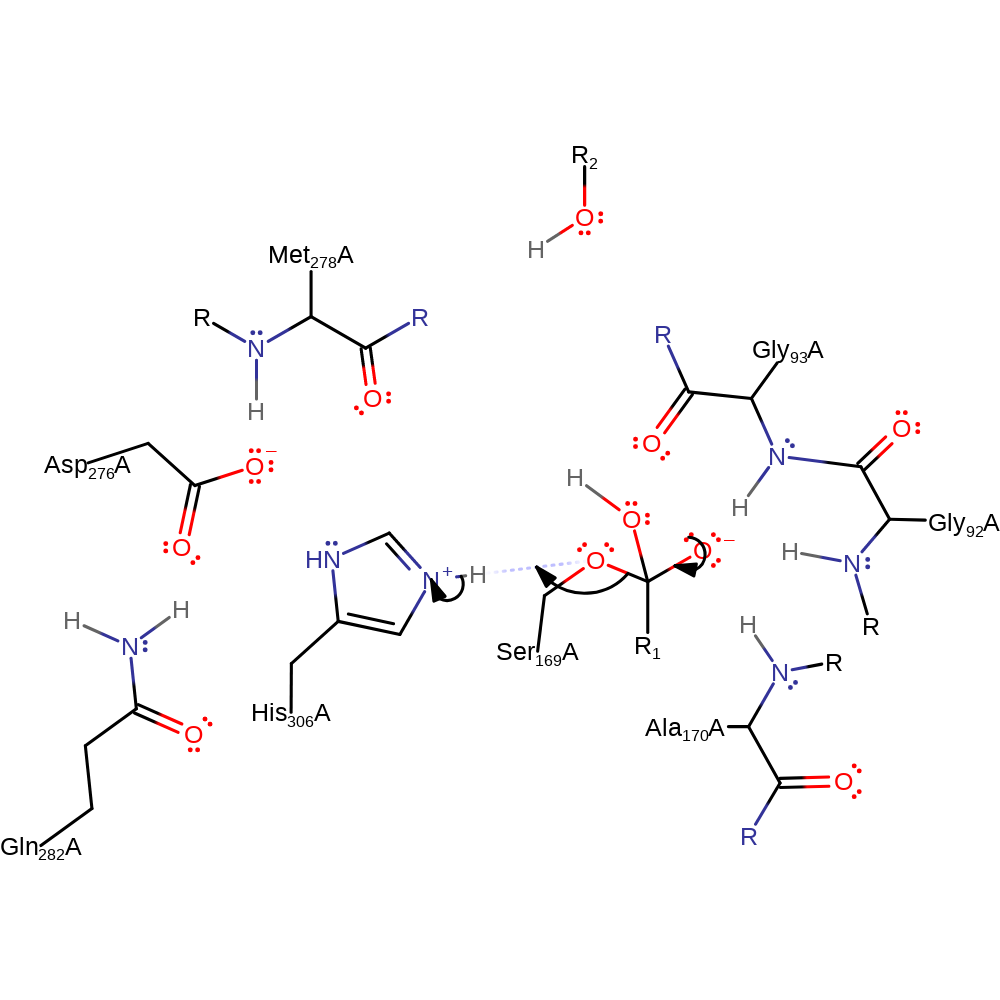
Step 4. Ser169 disconnects from the acyl complex and the active site is regenerated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser169A | hydrogen bond acceptor |
| Gly92A (main-N) | hydrogen bond donor |
| Gly93A (main-N) | hydrogen bond donor |
| Ala170A (main-N) | hydrogen bond donor |
| His306A | hydrogen bond donor |
| Gln282A | hydrogen bond donor |
| Met278A (main-N) | hydrogen bond donor |
| Asp276A | hydrogen bond acceptor |
| His306A | promote heterolysis |
| Ser169A | nucleofuge |





 Download:
Download: