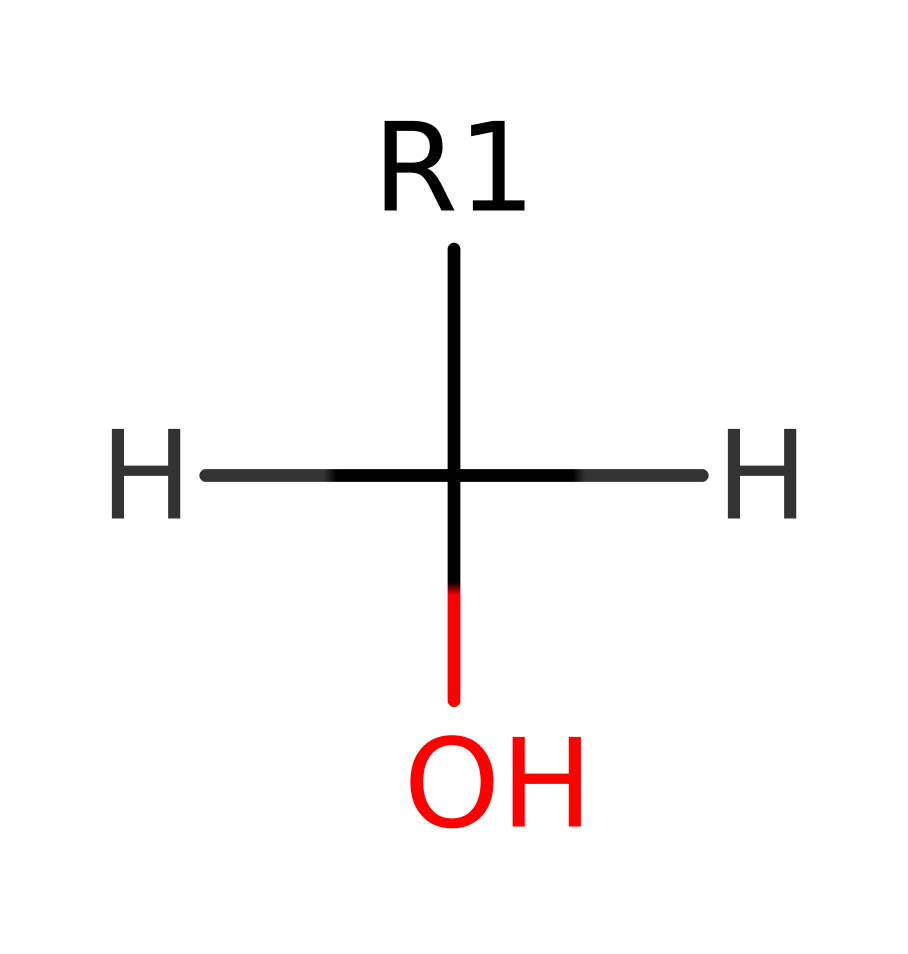
Haloalkane dehalogenase (subfamily 1)
Catalyzes hydrolytic cleavage of carbon-halogen bonds in halogenated aliphatic compounds, leading to the formation of the corresponding primary alcohols, halide ions and protons. Has a broad substrate specificity, which includes terminally mono- and di- chlorinated and brominated alkanes (up to C4 only). The highest activity was found with 1,2-dichloroethane, 1,3-dichloropropane, and 1,2-dibromoethane.
Reference Protein and Structure
- Sequence
-
P22643
 (3.8.1.5)
(3.8.1.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Xanthobacter autotrophicus (Bacteria)

- PDB
-
1b6g
- HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING CHLORIDE
(1.15 Å)



- Catalytic CATH Domains
-
3.40.50.1820
 (see all for 1b6g)
(see all for 1b6g)
Enzyme Reaction (EC:3.8.1.5)
Enzyme Mechanism
Introduction
Asp124 (activated through an Asp-His-HOH-Asp charge relay system) initiates a nucleophilic attack on the halide substrate, forming a covalent alkyl-enzyme intermediate with the halide ion being eliminated in an Sn2 reaction.
The water in the Asp-His-HOH-Asp charge relay system is activated by the histidine, and initiates the cleavage of the alkyl-enzyme intermediate. It is thought that the halide remains in the active site until the reaction is complete.
Catalytic Residues Roles
| UniProt | PDB* (1b6g) | ||
| Asp124 | Asp124A | Acts as the catalytic nucleophile. | covalent catalysis |
| Asp260 | Asp260A | Activates the catalytic nucleophile through its interaction with the second residue of the triad (histidine). | activator, electrostatic stabiliser |
| His289 | His289A | Activates the nucleophilic aspartate and acts as a general acid/base to activate the catalytic water molecule. | proton shuttle (general acid/base) |
| Trp125, Trp175 | Trp125A, Trp175A | Act to stabilise the halide ion. | electrostatic stabiliser |
Chemical Components
References
- Pavlová M et al. (2007), J Struct Biol, 157, 384-392. The identification of catalytic pentad in the haloalkane dehalogenase DhmA from Mycobacterium avium N85: Reaction mechanism and molecular evolution. DOI:10.1016/j.jsb.2006.09.004. PMID:17084094.
- Zhang Y et al. (2012), Int J Quantum Chem, 112, 889-899. Effect of electrostatic interaction on the mechanism of dehalogenation catalyzed by haloalkane dehalogenase. DOI:10.1002/qua.22509.
- Silberstein M et al. (2007), Biochemistry, 46, 9239-9249. Exploring the Binding Sites of the Haloalkane Dehalogenase DhlA fromXanthobacter autotrophicusGJ10†. DOI:10.1021/bi700336y. PMID:17645312.
- Lau EY et al. (2000), Proc Natl Acad Sci U S A, 97, 9937-9942. The importance of reactant positioning in enzyme catalysis: A hybrid quantum mechanics/molecular mechanics study of a haloalkane dehalogenase. DOI:10.1073/pnas.97.18.9937.
- Ridder IS et al. (1999), Acta Crystallogr D Biol Crystallogr, 55, 1273-1290. Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 Å resolution. DOI:10.1107/s090744499900534x. PMID:10393294.
- Pikkemaat MG et al. (1999), Biochemistry, 38, 12052-12061. Crystallographic and Kinetic Evidence of a Collision Complex Formed during Halide Import in Haloalkane Dehalogenase†. DOI:10.1021/bi990849w.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp124A | covalent catalysis |
| His289A | proton shuttle (general acid/base) |
| Asp260A | activator |
| Trp125A | electrostatic stabiliser |
| Trp175A | electrostatic stabiliser |
| Asp260A | electrostatic stabiliser |