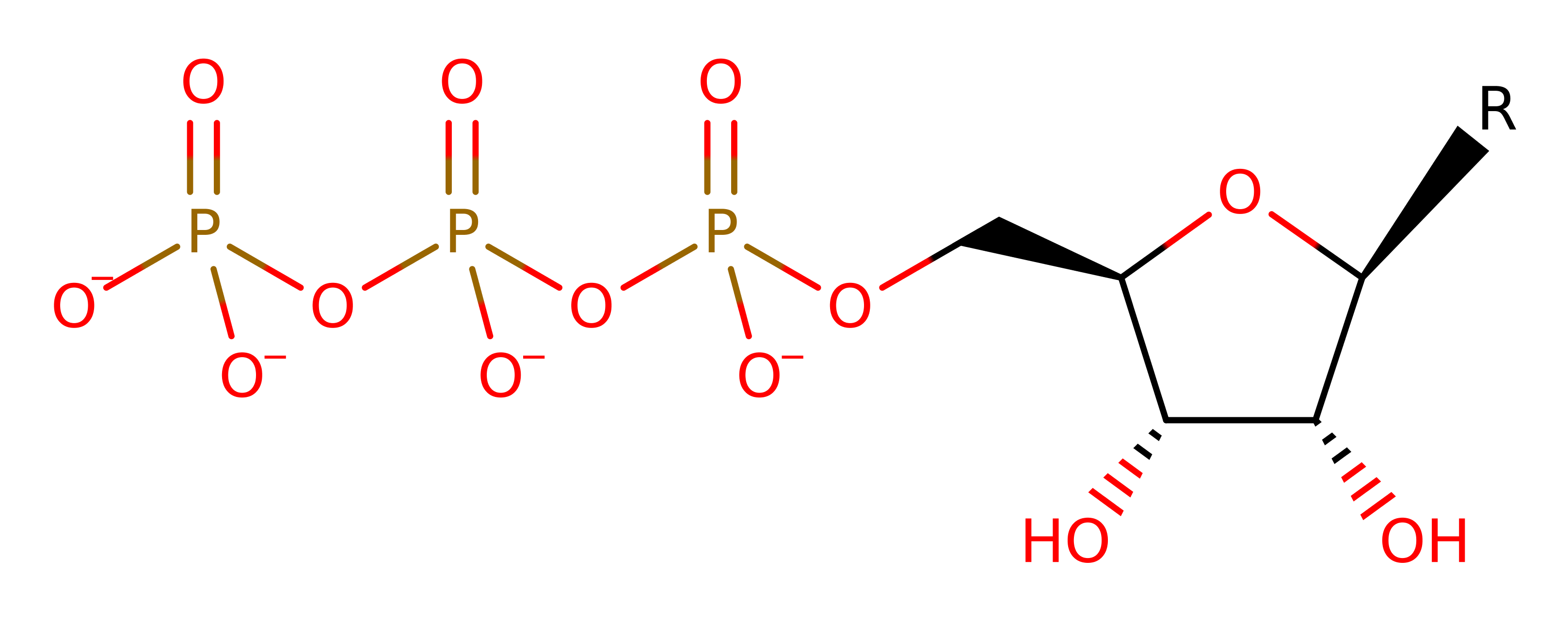
Ribonucleoside-triphosphate reductase (class III)
Ribonucleotide reductases (RNRs) catalyse the reductive synthesis of deoxyribonucleotides from their corresponding ribonucleotides. Providing the precursors necessary for DNA synthesis. This is a class III (anaerobic) RNR. The mechanism of the enzyme involves a glycine-centred radical (which is formed by the radical SAM superfamily protein NrdG).
Class III RNRs, found in strict or facultative anaerobic bacteria, bacteriophage, and archaea, use an FeS cluster and S-adenosylmethionine to generate a glycyl radical. The class III enzyme from phage T4 consists of two subunits, the larger of which contains the active and allosteric sites.
The protein binds a zinc ion in a position remote from the active site which has been shown to be critical for function. It is thought that this metal ion is responsible for protecting the enzyme from loss of its ground state radical.
Reference Protein and Structure
- Sequence
-
P07071
 (1.1.98.6)
(1.1.98.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Enterobacteria phage T4 (Virus)

- PDB
-
1h7a
- Structural basis for allosteric substrate specificity regulation in class III ribonucleotide reductases: NRDD in complex with dATP
(2.75 Å)



- Catalytic CATH Domains
-
3.20.70.20
 (see all for 1h7a)
(see all for 1h7a)
Enzyme Reaction (EC:1.17.4.2)
Enzyme Mechanism
Introduction
Upon substrate binding, the radical is transferred from G580 to the substrate via a first cysteine residue (C290) to yield a substrate radical, which is then reduced by formate via another essential active-site cysteine residue (C79) to give a deoxyribonucleotide radical. The latter is finally reduced to the deoxyribonucleotide product with concomitant recycling of the glycyl radical.
Catalytic Residues Roles
| UniProt | PDB* (1h7a) | ||
| Gly580 | Ala580A | It exists in the ground state as a glycyl radical. It is the initiating radical in the reaction. | hydrogen radical relay |
| Cys290, Cys79 | Cys290A, Cys79A | Act as hydrogen atom shuttles during the reaction. | hydrogen radical relay |
| Met288 | Met288A | Helps stabilise the radical formed on Cys79. | radical stabiliser |
| Asn311, Asn78 | Asn311A, Asn78A | Acts to stabilise and activate the formate substrate. | electrostatic stabiliser |
| Glu446, Ser292, Tyr441 | Glu446A, Ser292A, Tyr441A | Stabilises the intermediates. | electrostatic stabiliser |
Chemical Components
References
- Wei Y et al. (2014), J Am Chem Soc, 136, 9001-9013. A Chemically Competent Thiosulfuranyl Radical on theEscherichia coliClass III Ribonucleotide Reductase. DOI:10.1021/ja5030194. PMID:24827372.
- Luttringer F et al. (2009), J Biol Inorg Chem, 14, 923-933. The Zn center of the anaerobic ribonucleotide reductase from E. coli. DOI:10.1007/s00775-009-0505-9. PMID:19381696.
- Cho K et al. (2004), J Phys Chem B, 108, 2056-2065. Density Functional Calculations on Class III Ribonucleotide Reductase: Substrate Reaction Mechanism with Two Formates. DOI:10.1021/jp035280u.
- Fontecave M et al. (2002), Prog Nucleic Acid Res Mol Biol, 95-127. Deoxyribonucleotide synthesis in anaerobic microorganisms: The class III ribonucleotide reductase. DOI:10.1016/s0079-6603(02)72068-0.
- Andersson J et al. (2001), J Biol Chem, 276, 40457-40463. Two Active Site Asparagines Are Essential for the Reaction Mechanism of the Class III Anaerobic Ribonucleotide Reductase from Bacteriophage T4. DOI:10.1074/jbc.m106863200. PMID:11526118.
- Cho K et al. (2001), J Phys Chem B, 105, 6445-6452. The Substrate Reaction Mechanism of Class III Anaerobic Ribonucleotide Reductase. DOI:10.1021/jp0107614.
- Logan DT (1999), Science, 283, 1499-1504. A Glycyl Radical Site in the Crystal Structure of a Class III Ribonucleotide Reductase. DOI:10.1126/science.283.5407.1499.
- Stubbe J et al. (1998), Chem Rev, 98, 705-762. Protein Radicals in Enzyme Catalysis. DOI:10.1021/cr9400875. PMID:11848913.
- Mulliez E et al. (1995), Proc Natl Acad Sci U S A, 92, 8759-8762. Formate is the hydrogen donor for the anaerobic ribonucleotide reductase from Escherichia coli. PMID:7568012.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala580A | hydrogen radical relay |
| Cys79A | hydrogen radical relay |
| Met288A | radical stabiliser |
| Cys290A | hydrogen radical relay |
| Ser292A | electrostatic stabiliser |
| Asn311A | electrostatic stabiliser |
| Tyr441A | electrostatic stabiliser |
| Glu446A | electrostatic stabiliser |
| Asn78A | activator, electrostatic stabiliser |