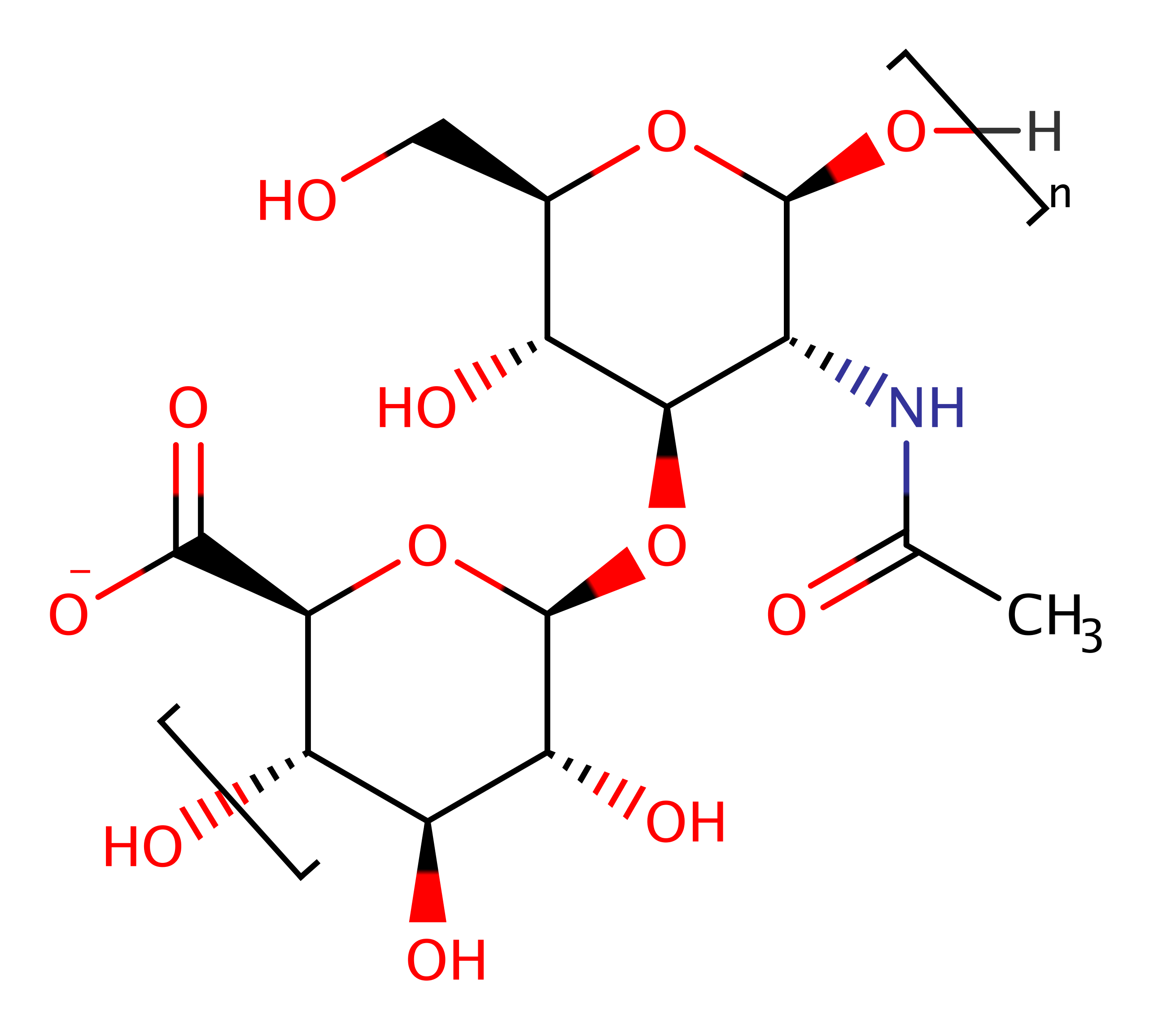
Hyaluronate lyase
Bacterial hyaluronan lyases (Hyal) belong to the polysaccharide lyase 8 family and degrade hyaluronan. Hyaluronan is an important component of the extracellular matrix, and involved in microbial spread. Hyal inhibitors may serve as tools to study the role of the enzyme, its substrates and products in the course of bacterial infections. Moreover, such enzyme inhibitors are potential candidates for antibacterial combination therapy.
Hyal catalyse the degradation of hyaluronan by a beta-elimination reaction. Also acts on chondroitin. Ultimately, they break the polysaccharide down to its constituent parts, more systematically known as 3-(4-deoxy-alpha-L-threo-hex-4-enopyranosyluronic acid)-2-acetamido-2-deoxy-D-glucose.
Reference Protein and Structure
- Sequence
-
Q54873
 (4.2.2.1)
(4.2.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Streptococcus pneumoniae TIGR4 (Bacteria)

- PDB
-
1c82
- MECHANISM OF HYALURONAN BINDING AND DEGRADATION: STRUCTURE OF STREPTOCOCCUS PNEUMONIAE HYALURONATE LYASE IN COMPLEX WITH HYALURONIC ACID DISACCHARIDE AT 1.7 A RESOLUTION
(1.7 Å)



- Catalytic CATH Domains
-
1.50.10.100
 (see all for 1c82)
(see all for 1c82)
Enzyme Reaction (EC:4.2.2.1)
Enzyme Mechanism
Introduction
Binding of the enzyme utilises its largely positive cleft to the negatively charged hyaluronan substrate and the precise relative positioning of HA and the enzyme by the hydrophobic patch residues Trp371, Trp372, and Phe423 interacting with hydrophobic sugar rings of HA.
In the active center, Asn349 attracts electrons on the carboxylate group of glucuronic acid, making the C5 proton more acidic. This is followed by the removal of a relatively acidic C5 proton by the Tyr408 side chain, which is negatively charged in the enzyme ground state thoruhg its interactions with His399 and Arg362. This results in the formation of an unsaturated C4–C5 bond and elimination of the product which abstracts a proton from Tyr408, regenerating the active site.
The consecutive release of the generated disaccharide product from the cleft uses the electrostatic repulsion of the negative product from the residues of the negative patch Glu468, Asp478, and Thr480. The final step is the translocation of the remaining truncated HA substrate in the cleft by one disaccharide unit in the direction of the reducing end of HA.
Catalytic Residues Roles
| UniProt | PDB* (1c82) | ||
| His516 | His399(232)A | Stabilises the intermediates. | modifies pKa |
| Tyr525 | Tyr408(241)A | Abstracts the acidic, axially positioned C5 proton, resulting in the formation of an unsaturated C4–C5 bond on glucuronic acid. | proton shuttle (general acid/base) |
| Asn466 | Asn349(182)A | Asn349 attracts electrons on the carboxylate group of glucuronic acid, making the C5 proton more acidic. | modifies pKa |
| Arg579 | Arg462(295)A | Facilitates the formation of the initial enzyme-substrate complex via the proton transfer from Y408 to H399. Positive charge ensures that Y408 is negatively charged and H399 positively charged. | modifies pKa |
Chemical Components
References
- Li S et al. (2000), EMBO J, 19, 1228-1240. Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. DOI:10.1093/emboj/19.6.1228. PMID:10716923.
- Li F et al. (2015), J Mol Model, 21, 196-. Functional role of R462 in the degradation of hyaluronan catalyzed by hyaluronate lyase from Streptococcus pneumoniae. DOI:10.1007/s00894-015-2724-z. PMID:26169310.
- Moore KB 3rd et al. (2014), Chemistry, 20, 990-998. Streptococcal Hyaluronate Lyase Reveals the Presence of a Structurally Significant CH⋅⋅⋅O Hydrogen Bond. DOI:10.1002/chem.201303231. PMID:24375830.
- Suits MD et al. (2014), J Biol Chem, 289, 27264-27277. Conformational Analysis of the Streptococcus pneumoniae Hyaluronate Lyase and Characterization of Its Hyaluronan-specific Carbohydrate-binding Module. DOI:10.1074/jbc.m114.578435. PMID:25100731.
- Zheng M et al. (2013), J Phys Chem B, 117, 10161-10172. Catalytic Mechanism of Hyaluronate Lyase fromSpectrococcus pneumonia: Quantum Mechanical/Molecular Mechanical and Density Functional Theory Studies. DOI:10.1021/jp406206s. PMID:23944739.
- Zheng M et al. (2012), J Phys Chem B, 116, 11166-11172. Initial Events in the Degradation of Hyaluronan Catalyzed by Hyaluronate Lyase from Spectrococcus pneumoniae: QM/MM Simulation. DOI:10.1021/jp306754a. PMID:22916709.
- Braun S et al. (2011), Eur J Med Chem, 46, 4419-4429. Design of benzimidazole- and benzoxazole-2-thione derivatives as inhibitors of bacterial hyaluronan lyase. DOI:10.1016/j.ejmech.2011.07.014. PMID:21803461.
- Joshi HV et al. (2009), Proteins, 76, 30-46. Domain motions of hyaluronan lyase underlying processive hyaluronan translocation. DOI:10.1002/prot.22316. PMID:19089975.
- Akhtar MS et al. (2006), J Biol Chem, 281, 28336-28344. Insights into the Mechanism of Action of Hyaluronate Lyase: ROLE OF C-TERMINAL DOMAIN AND Ca2+ IN THE FUNCTIONAL REGULATION OF ENZYME. DOI:10.1074/jbc.m601165200. PMID:16854993.
- Rigden DJ et al. (2003), J Biol Chem, 278, 50596-50606. Structures of Streptococcus pneumoniae Hyaluronate Lyase in Complex with Chondroitin and Chondroitin Sulfate Disaccharides: INSIGHTS INTO SPECIFICITY AND MECHANISM OF ACTION. DOI:10.1074/jbc.m307596200. PMID:14523022.
- Akhtar MS et al. (2003), J Biol Chem, 278, 25509-25516. Streptococcus pneumoniae Hyaluronate Lyase Contains Two Non-cooperative Independent Folding/Unfolding Structural Domains: CHARACTERIZATION OF FUNCTIONAL DOMAIN AND INHIBITORS OF ENZYME. DOI:10.1074/jbc.m301894200. PMID:12719417.
- Nukui M et al. (2003), J Biol Chem, 278, 3079-3088. The Function of Hydrophobic Residues in the Catalytic Cleft ofStreptococcus pneumoniae Hyaluronate Lyase: KINETIC CHARACTERIZATION OF MUTANT ENZYME FORMS. DOI:10.1074/jbc.m204999200. PMID:12446724.
- Mello LV et al. (2002), J Biol Chem, 277, 36678-36688. Structure and Flexibility of Streptococcus agalactiaeHyaluronate Lyase Complex with Its Substrate: INSIGHTS INTO THE MECHANISM OF PROCESSIVE DEGRADATION OF HYALURONAN. DOI:10.1074/jbc.m205140200. PMID:12130645.
- Jedrzejas MJ et al. (2002), J Biol Chem, 277, 28287-28297. Mechanism of Hyaluronan Degradation by Streptococcus pneumoniae Hyaluronate Lyase. STRUCTURES OF COMPLEXES WITH THE SUBSTRATE. DOI:10.1074/jbc.m112009200. PMID:11991948.
- Li S et al. (2001), J Biol Chem, 276, 41407-41416. Hyaluronan Binding and Degradation byStreptococcus agalactiae Hyaluronate Lyase. DOI:10.1074/jbc.m106634200. PMID:11527972.
- Kelly SJ et al. (2001), Glycobiology, 11, 297-304. Kinetic properties of Streptococcus pneumoniae hyaluronate lyase. DOI:10.1093/glycob/11.4.297.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn349(182)A | modifies pKa |
| His399(232)A | modifies pKa |
| Tyr408(241)A | proton shuttle (general acid/base) |
| Arg462(295)A | modifies pKa |