Cyclomaltodextrin glucanotransferase
Cyclodextrin glycosyltransferases (CGTases) are industrially important enzymes that produce cyclodextrins from starch by intramolecular transglycosylation. They belong to the important alpha-amylase family (family 13) of glycosyl hydrolases and catalyse the cyclisation of part of a (1->4)-alpha-D-glucan chain by the formation of a (1->4)-alpha-D-glucosidic bond. This enzyme also acts as a hydrolase or a transglycosylase, depending on the identity of the acceptor molecule in the final stages of the reaction.
Reference Protein and Structure
- Sequence
-
P43379
 (2.4.1.19)
(2.4.1.19)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus circulans (Bacteria)

- PDB
-
1cdg
- NUCLEOTIDE SEQUENCE AND X-RAY STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE FROM BACILLUS CIRCULANS STRAIN 251 IN A MALTOSE-DEPENDENT CRYSTAL FORM
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.80
 (see all for 1cdg)
(see all for 1cdg)
- Cofactors
- Calcium(2+) (3)
Enzyme Reaction (EC:2.4.1.19)
Enzyme Mechanism
Introduction
This enzyme has an Asp-Asp-Glu catalytic triad that is absolutely conserved in all the amylolytic enzymes.This triad occurs in the so-called domain A of the protein.
The first step of the cyclisation reaction is the binding of the starch chain at binding site 1 (in the E-domain, which includes part of the Ca3 binding site). After this event, binding is extended to the active site through a secondary binding site (also in the E domain) which is located near a groove leading to the active site.
Cleavage of the starch chain is initiated by the nucleophilic attack of Asp229 at one of the alpha(1-->4) glycosidic bonds (breaking the bond) leading to a beta(1-->4) glycosidically linked covalent intermediate. In the cyclase reaction, the non-reducing end of the covalently linked intermediate then migrates to the acceptor site. This acceptor then attacks the −1 glucose C1 atom leading to a cyclodextrin product (cyclisation).
If the acceptor is a water molecule, the the linear hydrolysis product is formed (hydrolysis).
If the acceptor molecule is another linear maltooligosaccharide then the product is a longer linear oligosaccharide (transglycosylation or disproportionation).
Catalytic Residues Roles
| UniProt | PDB* (1cdg) | ||
| Asp355 | Asp328A | Negatively charged side chain also stabilises the oxocarbenium transition states | hydrogen bond acceptor, electrostatic stabiliser |
| Asp256 | Asp229A | Acts as a nucleophile to attack the sugar, forming the covalent linkage within the intermediate. Its carboxylate group carbonyl oxygen also stabilises the transition state. | nucleofuge, nucleophile |
| Glu284 | Glu257A | Acts as a base protonates the glycosidic oxygen of the scissile bond in the first step and then deprotonates the attacking hydroxyl group in the second step. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Arg254, His354 | Arg227A, His327A | Helps stabilise the intermediates formed. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton transfer, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regenerated, intermediate terminated, intermediate collapseReferences
- Uitdehaag JC et al. (1999), Nat Struct Biol, 6, 432-436. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the alpha-amylase family. DOI:10.1038/8235. PMID:10331869.
- Li Z et al. (2016), Int J Biol Macromol, 83, 111-116. Asp577 mutations enhance the catalytic efficiency of cyclodextrin glycosyltransferase from Bacillus circulans. DOI:10.1016/j.ijbiomac.2015.11.042. PMID:26608005.
- Ban X et al. (2015), Int J Biol Macromol, 76, 224-229. Mutations at calcium binding site III in cyclodextrin glycosyltransferase improve β-cyclodextrin specificity. DOI:10.1016/j.ijbiomac.2015.02.036. PMID:25748847.
- Kumar V (2010), Carbohydr Res, 345, 893-898. Analysis of the key active subsites of glycoside hydrolase 13 family members. DOI:10.1016/j.carres.2010.02.007. PMID:20227065.
- van der Veen BA et al. (2000), Eur J Biochem, 267, 658-665. The three transglycosylation reactions catalyzed by cyclodextrin glycosyltransferase from Bacillus circulans (strain 251) proceed via different kinetic mechanisms. DOI:10.1046/j.1432-1327.2000.01031.x.
- Uitdehaag JCM et al. (2000), Biochemistry, 39, 7772-7780. Structures of Maltohexaose and Maltoheptaose Bound at the Donor Sites of Cyclodextrin Glycosyltransferase Give Insight into the Mechanisms of Transglycosylation Activity and Cyclodextrin Size Specificity†,‡. DOI:10.1021/bi000340x.
- Mosi R et al. (1997), Biochemistry, 36, 9927-9934. Trapping and Characterization of the Reaction Intermediate in Cyclodextrin Glycosyltransferase by Use of Activated Substrates and a Mutant Enzyme†. DOI:10.1021/bi970618u. PMID:9245426.
- Knegtel RM et al. (1995), J Biol Chem, 270, 29256-29264. Crystallographic Studies of the Interaction of Cyclodextrin Glycosyltransferase from Bacillus circulans Strain 251 with Natural Substrates and Products. DOI:10.1074/jbc.270.49.29256. PMID:7493956.
- Lawson CL et al. (1994), J Mol Biol, 236, 590-600. Nucleotide Sequence and X-ray Structure of Cyclodextrin Glycosyltransferase from Bacillus circulans Strain 251 in a Maltose-dependent Crystal Form. DOI:10.1006/jmbi.1994.1168. PMID:8107143.
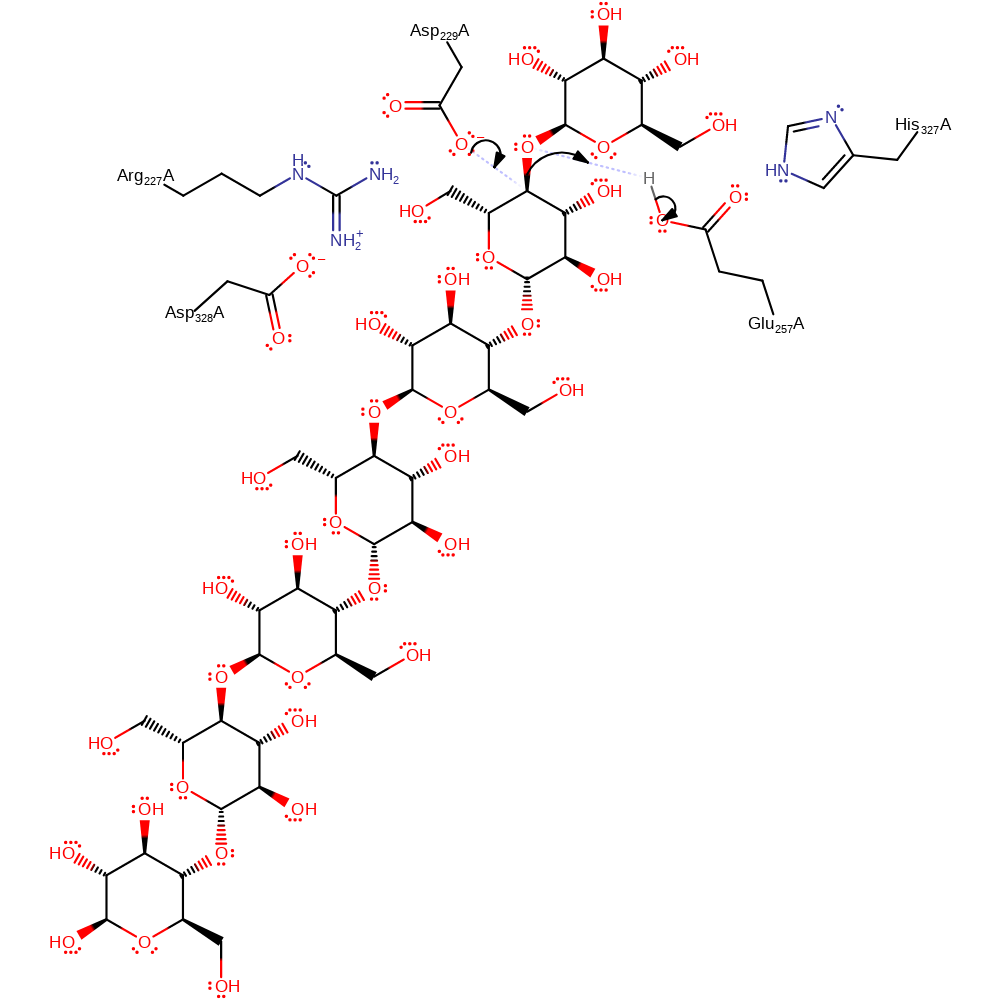
Step 1. Asp229 initiates a nucleophilic attack upon the carbon of the ether linkage between the two sugar rings in a substitution reaction, which eliminates a free sugar molecule, with concomitant deprotonation of Glu257.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg227A | hydrogen bond donor, electrostatic stabiliser |
| His327A | hydrogen bond donor, electrostatic stabiliser |
| Asp328A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu257A | hydrogen bond donor |
| Glu257A | proton donor |
| Asp229A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton transfer
Step 2. Glu257 deprotonates a the acceptor OH, which initiates a nucleophilic attack upon the covalently bound sugar in a substitution reaction, eliminating Asp229.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg227A | hydrogen bond donor, electrostatic stabiliser |
| His327A | hydrogen bond donor, electrostatic stabiliser |
| Asp328A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu257A | hydrogen bond acceptor, proton acceptor |
| Asp229A | nucleofuge |


 Download:
Download: