Site-specific DNA-methyltransferase (adenine-specific)
N-6 adenine-specific DNA-methyltransferases catalyse the transfer of a methyl group from S-adenosyl-L-methionine to either the exocyclic N-6 of an adenine or to N-4 of a cytosine contained in a specific DNA recognition sequence. Methylation protects the DNA from restriction from host endonucleases.
Reference Protein and Structure
- Sequence
-
P14385
 (2.1.1.72)
(2.1.1.72)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermus aquaticus (Bacteria)

- PDB
-
2adm
- ADENINE-N6-DNA-METHYLTRANSFERASE TAQI
(2.6 Å)



- Catalytic CATH Domains
-
3.40.50.150
 (see all for 2adm)
(see all for 2adm)
Enzyme Reaction (EC:2.1.1.72)
Enzyme Mechanism
Introduction
The binding site for S-adenosyl-L-methionine is located in a cavity in the N-terminal domain. Its positive charge is compensated for by seven negatively charged residues, Glu22, Glu45, Glu71, Glu78, Glu84, Asp89 and Glu113. Hydrogen bonds occur with Asp89, Phe90, Glu71, Glu45, Glu22 and Thr23 in addition to many hydrophobic interactions. The DNA binds in the cleft formed by the N- and C-terminal domains which has a high positive electrostatic potential allowing interaction with the negative DNA phosphate backbone. The adenine to be methylated is brought to the active methyl group in S-adenosyl-L-methionine by looping out of the base into a gap between the flexible N-terminal loop and beta strand.
Since no basic residue is found in the vicinity of the 6-amino group, a catalytic mechanism in which N6 is activated by deprotonation can be ruled out. Instead, the amine is activated by interactions with Asn105 and the backbone carbonyl of Pro106. The reaction proceeds by a the nucleophilic attack of the adenine N6 on the methyl of SAM in an SN2-type reaction.
Catalytic Residues Roles
| UniProt | PDB* (2adm) | ||
| Asn105 | Asn105A | The observed hydrogen bonding of the 6-amino group with OD1 of Asn 105 should increase the electron density of N6 and contribute to its activation for nucleophilic attack. In addition, both hydrogen bond acceptors lie below the plane of the adenine ring and could pull the hydrogen atoms of the 6-amino group out of a coplanar arrangement with the attached purine ring. This could assist a hybridization change of N6 from sp2 towards sp3, where the lone pair of N6 is no longer conjugated with the aromatic system and, thus, should have a strongly enhanced nucleophilicity. Such a hybridization change would also place the lone pair at N6 in an ideal geometry for an inline attack on the activated methyl group of AdoMet | activator, hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Pro106 (main-C) | Pro106A (main-C) | The observed hydrogen bonding of the 6-amino group with the backbone oxygen of Pro 106 should increase the electron density of N6 and contribute to its activation for nucleophilic attack. In addition, both hydrogen bond acceptors lie below the plane of the adenine ring and could pull the hydrogen atoms of the 6-amino group out of a coplanar arrangement with the attached purine ring. This could assist a hybridization change of N6 from sp2 towards sp3, where the lone pair of N6 is no longer conjugated with the aromatic system and, thus, should have a strongly enhanced nucleophilicity. Such a hybridization change would also place the lone pair at N6 in an ideal geometry for an inline attack on the activated methyl group of AdoMet | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Tyr108, Phe196 | Tyr108A, Phe196A | Help bind and position the substrate for catalysis. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation, proton transfer, intermediate terminatedReferences
- Newby ZE et al. (2002), Proc Natl Acad Sci U S A, 99, 7922-7927. A theoretical examination of the factors controlling the catalytic efficiency of the DNA-(adenine-N6)-methyltransferase from Thermus aquaticus. DOI:10.1073/pnas.122231499. PMID:12060740.
- Goedecke K et al. (2001), Nat Struct Biol, 8, 121-125. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. DOI:10.1038/84104. PMID:11175899.
- Scavetta RD et al. (2000), Nucleic Acids Res, 28, 3950-3961. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. PMID:11024175.
- Pues H et al. (1999), Biochemistry, 38, 1426-1434. Functional Roles of the Conserved Aromatic Amino Acid Residues at Position 108 (Motif IV) and Position 196 (Motif VIII) in Base Flipping and Catalysis by the N6-Adenine DNA Methyltransferase fromThermus aquaticus†. DOI:10.1021/bi9818016. PMID:9931007.
- Schluckebier G et al. (1995), Gene, 157, 131-134. A model for DNA binding and enzyme action derived from crystallographic studies of the TaqI N6-adenine-methyltransferase. DOI:10.1016/0378-1119(94)00690-t. PMID:7607476.

Step 1. The 6-amino group of DNA adenine initiates a nucleophilic attack upon the methyl carbon of S-adenosyl-L-methionine in a substitution reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Pro106A (main-C) | hydrogen bond acceptor, activator |
| Asn105A | hydrogen bond acceptor, hydrogen bond donor, activator |
| Tyr108A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Phe196A | steric role |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation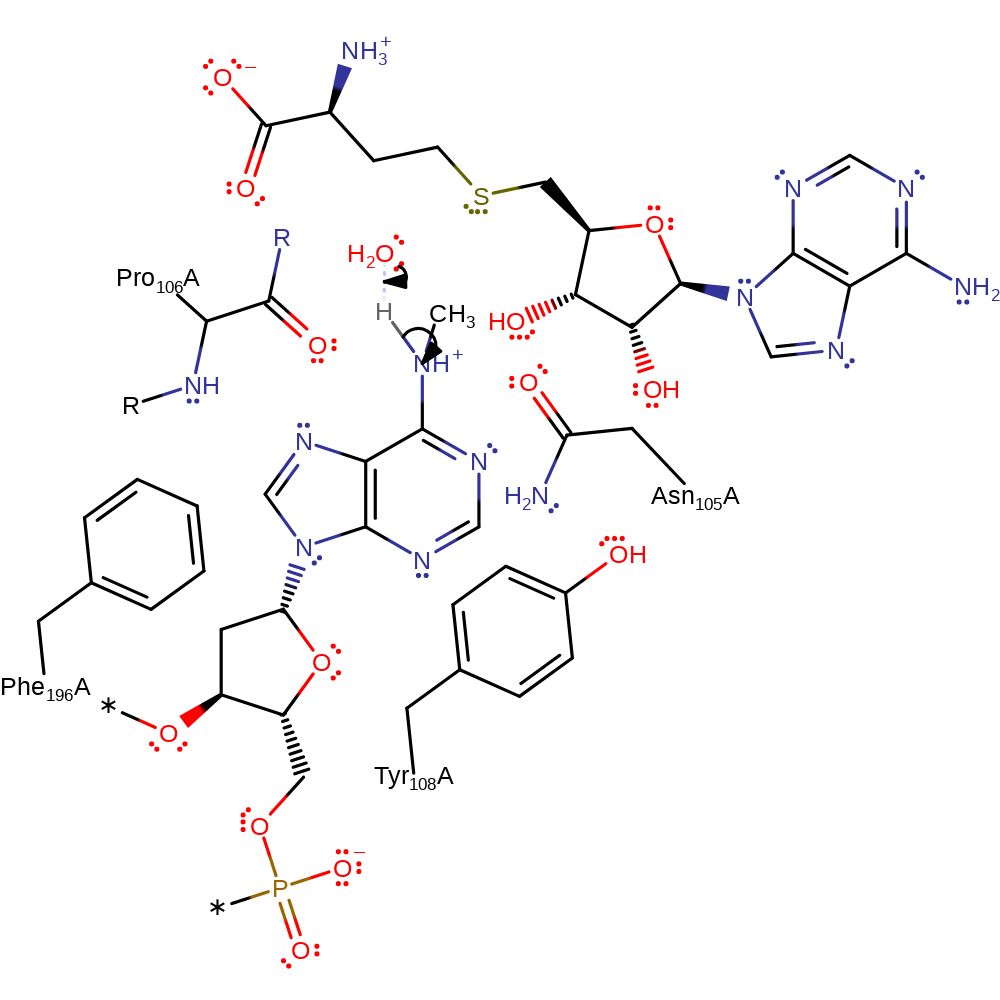
Step 2. Water deprotonates the 6-amino group of the DNA methyladenine product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn105A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Tyr108A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Pro106A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Phe196A | steric role |





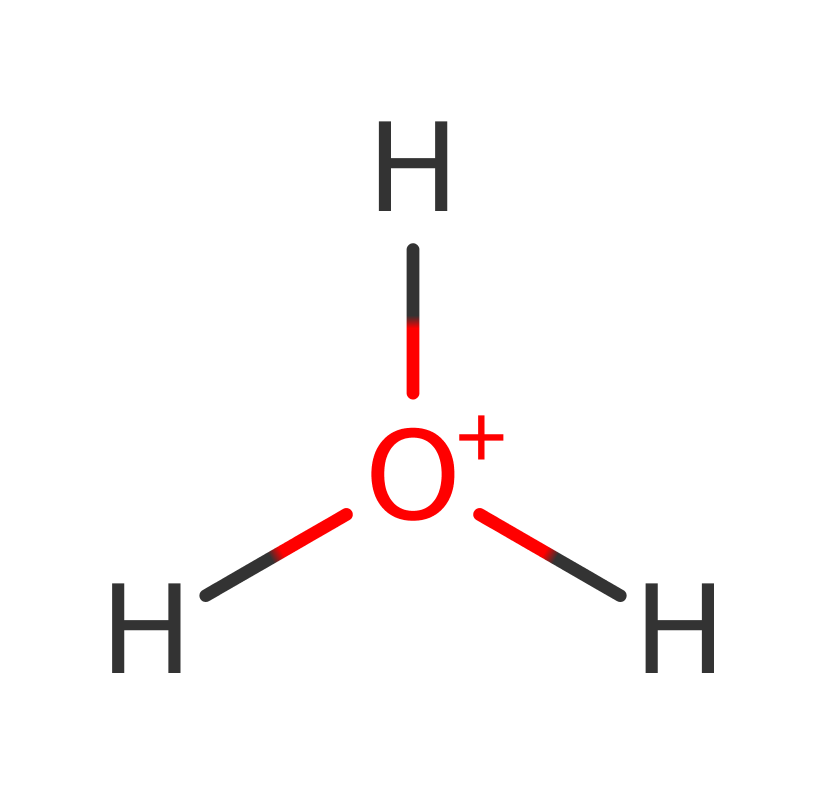
 Download:
Download: