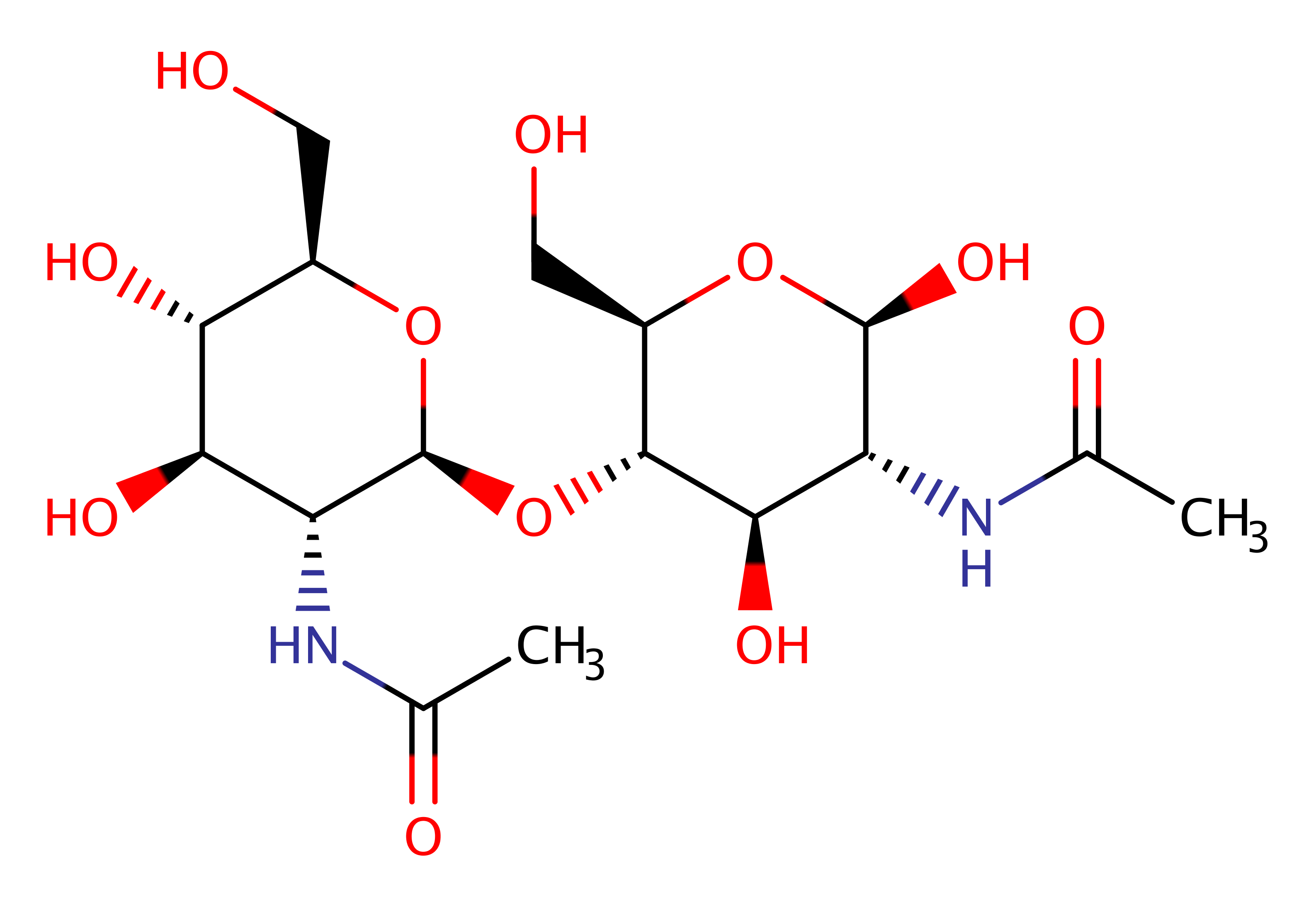
Chitinase (GH18, Class II)
Chitinase binds to chitin and randomly cleaves glycosidic linkages in chitin and chitodextrins in a non-processive mode, generating chitooligosaccharides and free ends on which exo-chitinases and exo-chitodextrinases can act. Chitin is a bio-polymer composed of 1,4-beta linked N-acetylglucosamine units, a major component of fungal cell walls and insect exoskeletons.
The bacterial chitinase enzyme is an 18-family member, and is used by bacteria to catalyse the degradation of chitin to form products which are then used as a source of energy.
Reference Protein and Structure
- Sequence
-
P07254
 (3.2.1.14)
(3.2.1.14)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Serratia marcescens (Bacteria)

- PDB
-
1ctn
- CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION
(2.3 Å)



- Catalytic CATH Domains
-
3.20.20.80
 (see all for 1ctn)
(see all for 1ctn)
Enzyme Reaction (EC:3.2.1.14)
Enzyme Mechanism
Introduction
Recent studies have shown the hydrolysis of 18-family chitinase to proceed through a substrate assisted mechanism with retention of configuration in the product. A putative oxocarbonium ion intermediate is stabilised by the anchimeric assistance of the sugar N-acetyl group after the donation of a proton from the catalytic carboxylate to the departing N acetylglucosamine unit. The anionic glutamate activates a hydrolytic water by abstracting a proton, forming a hydroxide which kicks out the N-acyl group in a SN2 mechanism and forms the C1-OH group of the product. The active site is then restored.
Glu315 is protonated and poised to cleave the glycosidic linkage between −1 and +1 sugar residues. One water molecule is H-bonded to the phenol hydroxyl of Tyr390 and to the NH of the 2-N-acetyl group. The approach of acidic residue Asp313 toward Glu315 forces the acetamido group of the -1 sugar to rotate around its C2−N2 bond, thus displacing the water molecule into the vicinity of Glu315. A proton from the water molecule will be taken up by the gamma carboxylate of Glu315 and the remaining hydroxide ion will be taken up by the anomeric C1 atom of the -1 sugar residue. Once hydrolysis is completed, the monosaccharide departs from subsite -1, and the disaccharide remains bound at the enzyme subsites +1, +2.
Catalytic Residues Roles
| UniProt | PDB* (1ctn) | ||
| Tyr390 | Tyr390(367)A | Tyr214 thought to play an important role in destabilising intermediates formed during the enzyme-catalyzed reaction. | electrostatic destabiliser |
| Asp313 | Asp313(290)A | Asp313 that interacts with Asp311 flips to its alternative position where it interacts with Glu315 thus forcing the substrate acetamido group of -1 sugar to rotate around the C2−N2 bond. | steric role |
| Asp311 | Asp311(288)A | Thought to be in a neutral charge state in the active site. It plays a critical role in catalysis by electrostatically stabilising the transition state and oxazolinium ion intermediate formed in the glycosylation step of the reaction. | electrostatic stabiliser |
| Glu315 | Glu315(292)A | The residue acts as a general acid to the transient oxocarbonium transition state. The resulting carboxylate group then acts as a general base towards a hydrolytic water, initiating SN2 displacement of the anchimeric N-acetyl group. | proton shuttle (general acid/base) |
Chemical Components
References
- Papanikolau Y et al. (2001), Biochemistry, 40, 11338-11343. High Resolution Structural Analyses of Mutant Chitinase A Complexes with Substrates Provide New Insight into the Mechanism of Catalysis†,‡. DOI:10.1021/bi010505h. PMID:11560481.
- Meekrathok P et al. (2017), J Chem Inf Model, 57, 572-583. Investigation of Ionization Pattern of the Adjacent Acidic Residues in the DXDXE Motif of GH-18 Chitinases Using Theoretical pKa Calculations. DOI:10.1021/acs.jcim.6b00536. PMID:28230366.
- Jitonnom J et al. (2014), J Phys Chem B, 118, 4771-4783. QM/MM Free-Energy Simulations of Reaction inSerratia marcescensChitinase B Reveal the Protonation State of Asp142 and the Critical Role of Tyr214. DOI:10.1021/jp500652x. PMID:24730355.
- Papanikolau Y et al. (2003), Acta Crystallogr D Biol Crystallogr, 59, 400-403. De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin. PMID:12554965.
- Bokma E et al. (2002), Eur J Biochem, 269, 893-901. Expression and characterization of active site mutants of hevamine, a chitinase from the rubber treeHevea brasiliensis. DOI:10.1046/j.0014-2956.2001.02721.x. PMID:11846790.
- Fukamizo T (2000), Curr Protein Pept Sci, 1, 105-124. Chitinolytic Enzymes: Catalysis, Substrate Binding, and their Application. DOI:10.2174/1389203003381450. PMID:12369923.
- Lin FP et al. (1999), IUBMB Life, 48, 199-204. Site-Directed Mutagenesis of Asp313, Glu315, and Asp391 Residues in Chitinase of Aeromonas Caviae. DOI:10.1080/713803487. PMID:10794597.
- Brameld KA et al. (1998), J Am Chem Soc, 120, 3571-3580. Substrate Distortion to a Boat Conformation at Subsite −1 Is Critical in the Mechanism of Family 18 Chitinases. DOI:10.1021/ja972282h.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu315(292)A | proton shuttle (general acid/base) |
| Asp311(288)A | electrostatic stabiliser |
| Tyr390(367)A | electrostatic destabiliser |
| Asp313(290)A | steric role |