Hypoxanthine phosphoribosyltransferase
Converts guanine to guanosine monophosphate, and hypoxanthine to inosine monophosphate. Transfers the 5-phosphoribosyl group from 5-phosphoribosylpyrophosphate onto the purine. Plays a central role in the generation of purine nucleotides through the purine salvage pathway and is involved in the first step of the subpathway that synthesizes IMP from hypoxanthine.
Reference Protein and Structure
- Sequence
-
P00492
 (2.4.2.8)
(2.4.2.8)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1bzy
- HUMAN HGPRTASE WITH TRANSITION STATE INHIBITOR
(2.0 Å)



- Catalytic CATH Domains
-
3.40.50.2020
 (see all for 1bzy)
(see all for 1bzy)
- Cofactors
- Magnesium(2+) (2) Metal MACiE
Enzyme Reaction (EC:2.4.2.8)
Enzyme Mechanism
Introduction
In a nucleophilic bimolecular substitution, Asp137 activates the guanine for attach on the C2 of the substrate ribose, the reaction proceeds via a positively charged transition state.
Catalytic Residues Roles
| UniProt | PDB* (1bzy) | ||
| Asp138 | Asp137A | Acts as a general acid/base. It deprotonates the guanine that initiated the nucleophilic attack on the ribose ring. It has been suggested [PMID:11258886] that a general base may not be required, but a strong hydrogen bond with the N7 of the purine substrate provides sufficient transition-state stabilisation to permit relatively efficient catalysis. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Arg200 | Arg199A | Binds the phosphate group, may be involved in electrostatic stabilisation of this group. | electrostatic stabiliser |
| Phe187 | Phe186A | Stabilises the transition state by pi-cation interactions. | electrostatic stabiliser |
| Glu134, Asp135 | Glu133A, Asp134A | Help stabilise the positively charged ribooxocarbenium ion at the transition state. | attractive charge-charge interaction, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, dephosphorylation, proton transfer, rate-determining step, native state of enzyme regenerated, inferred reaction stepReferences
- Héroux A et al. (1999), Biochemistry, 38, 14495-14506. Crystal Structure ofToxoplasma gondiiHypoxanthine-Guanine Phosphoribosyltransferase with XMP, Pyrophosphate, and Two Mg2+Ions Bound: Insights into the Catalytic Mechanism†,‡. DOI:10.1021/bi990508i. PMID:10545171.
- Karnawat V et al. (2015), Chemphyschem, 16, 2172-2181. Differential Distortion of Purine Substrates by Human andPlasmodium falciparumHypoxanthine-Guanine Phosphoribosyltransferase to Catalyse the Formation of Mononucleotides. DOI:10.1002/cphc.201500084. PMID:25944719.
- Gasik Z et al. (2013), Curr Pharm Des, 19, 4226-4240. Resolving differences in substrate specificities between human and parasite phosphoribosyltransferases via analysis of functional groups of substrates and receptors. PMID:23170881.
- Canyuk B et al. (2001), Biochemistry, 40, 2754-2765. The Role for an Invariant Aspartic Acid in Hypoxanthine Phosphoribosyltransferases Is Examined Using Saturation Mutagenesis, Functional Analysis, and X-ray Crystallography†. DOI:10.1021/bi001195q. PMID:11258886.
- Héroux A et al. (2000), Structure, 8, 1309-1318. Substrate deformation in a hypoxanthine-guanine phosphoribosyltransferase ternary complex: the structural basis for catalysis. PMID:11188695.
- Shi W et al. (1999), Nat Struct Biol, 6, 588-593. The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor. DOI:10.1038/9376. PMID:10360366.
- Xu Y et al. (1998), Biochemistry, 37, 4114-4124. Catalysis in Human Hypoxanthine-Guanine Phosphoribosyltransferase: Asp 137 Acts as a General Acid/Base†. DOI:10.1021/bi972519m. PMID:9521733.
- Jardim A et al. (1997), J Biol Chem, 272, 8967-8973. The Conserved Serine-Tyrosine Dipeptide in Leishmania donovani Hypoxanthine-guanine Phosphoribosyltransferase Is Essential for Catalytic Activity. DOI:10.1074/jbc.272.14.8967. PMID:9083019.

Step 1. Asp137 deprotonates the guanine which initiates a nucleophilic attack upon the C2 of the ribose ring in a substitution reaction, eliminating diphosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp137A | hydrogen bond acceptor |
| Glu133A | attractive charge-charge interaction, electrostatic stabiliser |
| Asp134A | attractive charge-charge interaction, electrostatic stabiliser |
| Phe186A | electrostatic stabiliser |
| Arg199A | electrostatic stabiliser |
| Asp137A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, dephosphorylation, proton transfer, rate-determining step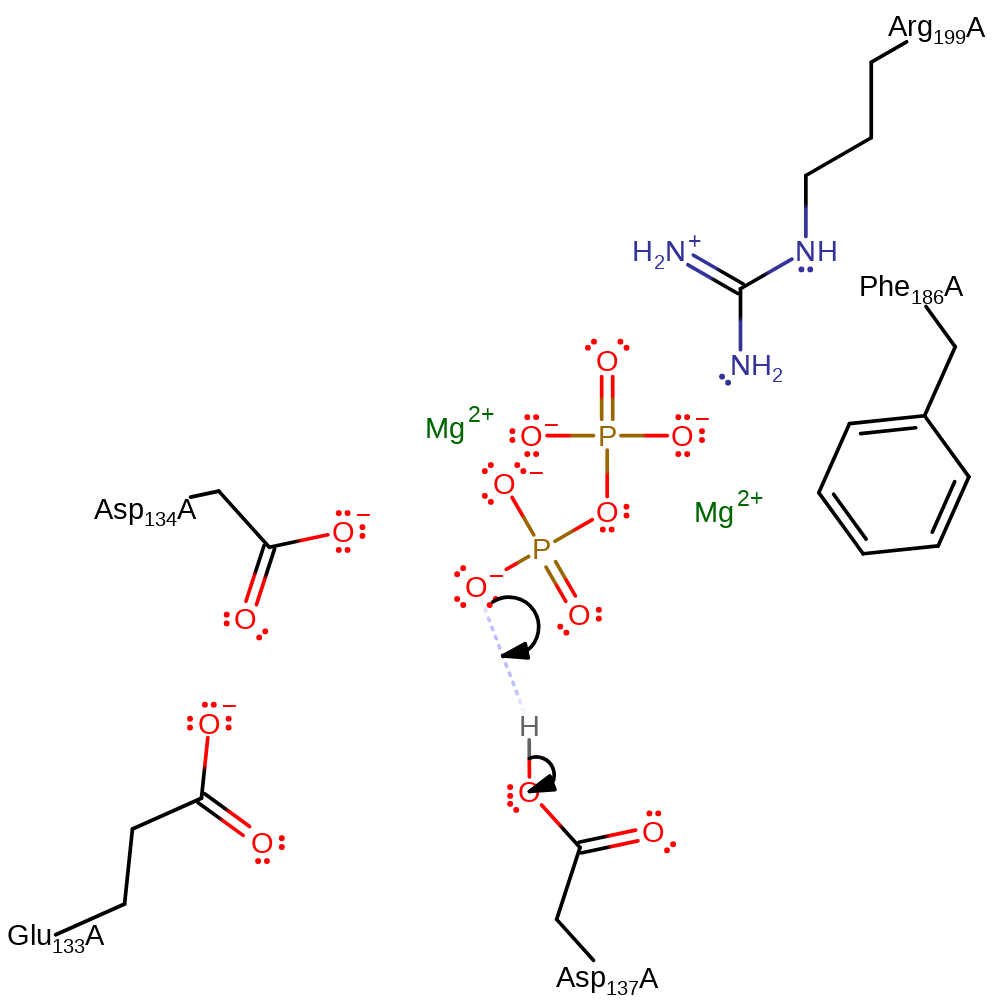
Step 2. The diphosphate product deprotonates Asp137 in an inferred return step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp137A | hydrogen bond donor, hydrogen bond acceptor, proton donor |



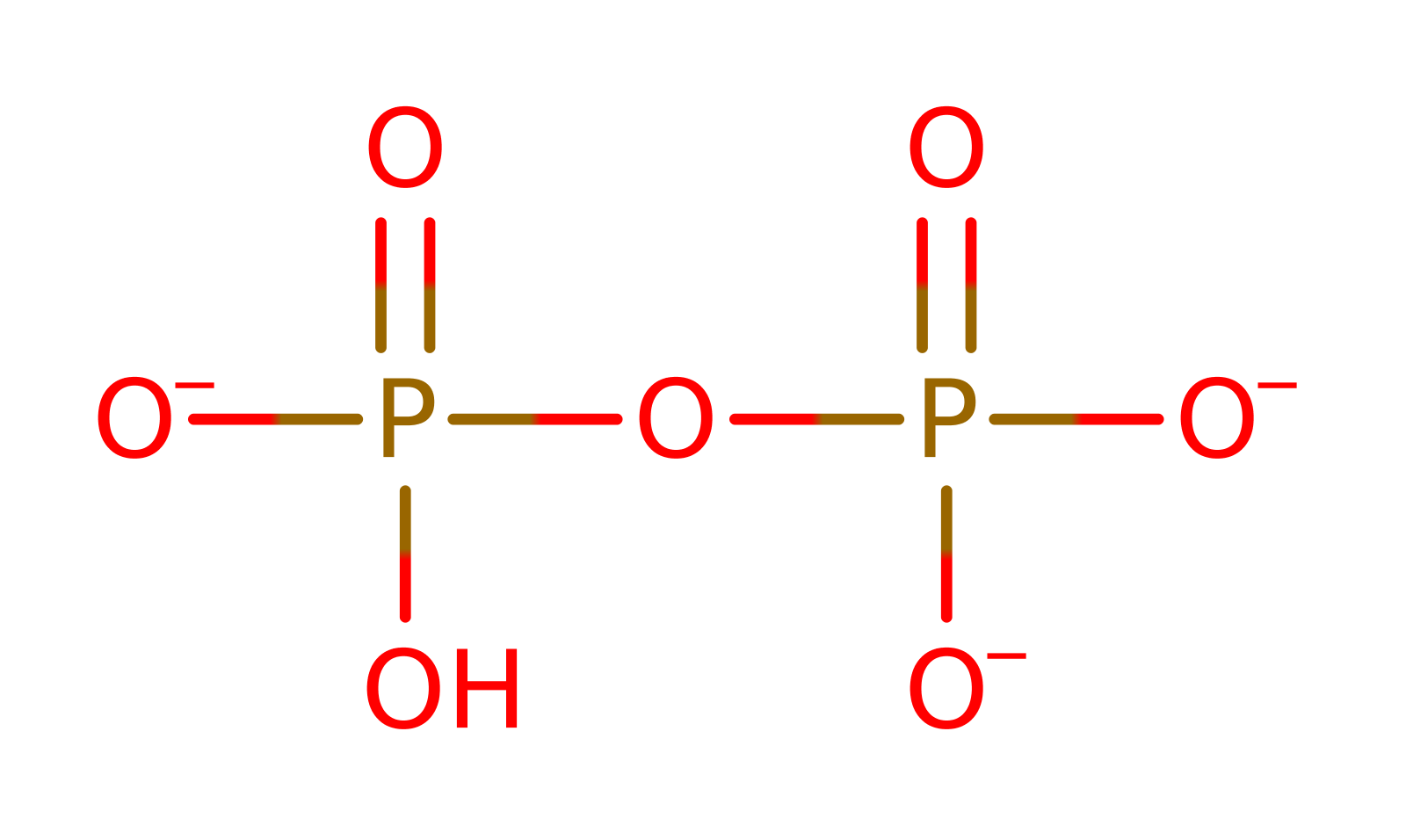
 Download:
Download: