Orotidine-5'-phosphate decarboxylase
Orotidine-5'-phosphate decarboxylase catalyses the decarboxylation of orotidine-5'-phosphate, the sixth and final step in the de novo pyrimidine biosynthesis of uridine monophosphate. It accelerates the reaction by 10^17 and hence is the most proficient enzyme discovered so far. In most prokaryotes the bioactive form is a homodimer. In higher organisms it is part of a bifunctional enzyme.
Reference Protein and Structure
- Sequence
-
P25971
 (4.1.1.23)
(4.1.1.23)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus subtilis subsp. subtilis str. 168 (Bacteria)

- PDB
-
1dbt
- CRYSTAL STRUCTURE OF OROTIDINE 5'-MONOPHOSPHATE DECARBOXYLASE FROM BACILLUS SUBTILIS COMPLEXED WITH UMP
(2.4 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1dbt)
(see all for 1dbt)
Enzyme Reaction (EC:4.1.1.23)
Enzyme Mechanism
Introduction
The enzyme has a novel mechanism: the carbanion generated by carbon dioxide loss is localised in an sp2 orbital perpendicular to the pi system of the pyrimidine. In all other decarboxylases, the carbanion is delocalised either into an adjacent carbonyl or into a covalently bound thiamin, pyridoxal or pyruvoyl cofactor.
The mechanism is a bimolecular electrophilic substitution SE2 in which decarboxylation and protonation occur in a step-wise manner. Firstly, the anionic carboxylate of the substrate is positioned in a negatively charged region close to the carboxylate of Asp60 and Asp65(B) and the carbon of the pyrimidine destined to become the carbanium is close to the positive protonated Lys62. This allows destabilisation of the ground state and the stabilisation of negative charge accumulation in the transition state.
Extensive hydrogen bonding interactions within the active site provide the binding energy required to force the carboxylate groups into close proximity. As the reaction proceeds, the weakly basic C-C bond linking the carboxy group to the pyrimidine becomes progressively more basic until proton transfer from the adjacent Lys62 occurs.
Catalytic Residues Roles
| UniProt | PDB* (1dbt) | ||
| Lys62 | Lys62A | The residue acts as a general acid to the cleaving carboxy-pyrimidine C-C bond in concert with decarboxylation. The residue is then reprotonated by solvent to regenerate the active site. Its pKa is modified through electrostatic interactions with the neighbouring carboxyl groups of Asp60 and Asp65(B). | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Arg215 | Arg215A | Binds the phosphate group, aids in stabilising the negatively charged transition state via long range interactions. | hydrogen bond donor, electrostatic destabiliser |
| Lys33 | Lys33A | Part of the catalytic tetrad. | hydrogen bond donor, electrostatic destabiliser, electrostatic stabiliser |
| Asp60, Asp65 | Asp60A, Asp65B | The carboxylate side chains remain deprotonated and anionic within close proximity to the substrate carboxyl group. Although the groups repel one another, the strength of the surrounding hydrogen bonding network directs them towards one another. These repulsive interactions increase the energy of the groundstate so it resembles that of the transition state and therefore reduces the reaction activation barrier while also stabilising the polar transition state. | repulsive charge-charge interaction, activator, hydrogen bond acceptor, electrostatic destabiliser |
| Thr123 (main-N) | Thr123A (main-N) | The backbone NH of Ser 127 (in M. thermoautotrophicus, Thr 123 here) increases as negative charge is delocalised to O4, this interaction could provide significant stabilisation of the intermediate. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
unimolecular elimination by the conjugate base, proton transfer, overall reactant used, overall product formed, decarboxylation, rate-determining step, native state of enzyme regeneratedReferences
- Appleby TC et al. (2000), Proc Natl Acad Sci U S A, 97, 2005-2010. The crystal structure and mechanism of orotidine 5'-monophosphate decarboxylase. DOI:10.1073/pnas.259441296. PMID:10681442.
- Jamshidi S et al. (2015), J Biomol Struct Dyn, 33, 404-417. Study of orotidine 5′-monophosphate decarboxylase in complex with the top three OMP, BMP, and PMP ligands by molecular dynamics simulation. DOI:10.1080/07391102.2014.881303. PMID:24559040.
- Goldman LM et al. (2014), J Am Chem Soc, 136, 10156-10165. Enzyme Architecture: Deconstruction of the Enzyme-Activating Phosphodianion Interactions of Orotidine 5′-Monophosphate Decarboxylase. DOI:10.1021/ja505037v. PMID:24958125.
- Amyes TL et al. (2012), Biochemistry, 51, 4630-4632. Orotidine 5′-Monophosphate Decarboxylase: Transition State Stabilization from Remote Protein–Phosphodianion Interactions. DOI:10.1021/bi300585e. PMID:22620855.
- Tsang WY et al. (2012), J Am Chem Soc, 134, 14580-14594. Proton Transfer from C-6 of Uridine 5′-Monophosphate Catalyzed by Orotidine 5′-Monophosphate Decarboxylase: Formation and Stability of a Vinyl Carbanion Intermediate and the Effect of a 5-Fluoro Substituent. DOI:10.1021/ja3058474. PMID:22812629.
- Desai BJ et al. (2012), Biochemistry, 51, 8665-8678. Conformational Changes in Orotidine 5′-Monophosphate Decarboxylase: A Structure-Based Explanation for How the 5′-Phosphate Group Activates the Enzyme. DOI:10.1021/bi301188k. PMID:23030629.
- Iiams V et al. (2011), Biochemistry, 50, 8497-8507. Mechanism of the Orotidine 5′-Monophosphate Decarboxylase-Catalyzed Reaction: Importance of Residues in the Orotate Binding Site. DOI:10.1021/bi2012355. PMID:21870810.
- Toth K et al. (2010), J Am Chem Soc, 132, 7018-7024. Product Deuterium Isotope Effects for Orotidine 5′-Monophosphate Decarboxylase: Effect of Changing Substrate and Enzyme Structure on the Partitioning of the Vinyl Carbanion Reaction Intermediate. DOI:10.1021/ja102408k. PMID:20441167.
- Thirumalairajan S et al. (2010), Chem Commun (Camb), 46, 3158-. Interrogation of the active site of OMP decarboxylase from Escherichia coli with a substrate analogue bearing an anionic group at C6. DOI:10.1039/b926894d. PMID:20424759.
- Heinrich D et al. (2009), Chemistry, 15, 6619-6625. Lys314 is a Nucleophile in Non-Classical Reactions of Orotidine-5′-Monophosphate Decarboxylase. DOI:10.1002/chem.200900397. PMID:19472232.
- Chan KK et al. (2009), Biochemistry, 48, 5518-5531. Mechanism of the Orotidine 5′-Monophosphate Decarboxylase-Catalyzed Reaction: Evidence for Substrate Destabilization,. DOI:10.1021/bi900623r. PMID:19435314.
- Amyes TL et al. (2008), J Am Chem Soc, 130, 1574-1575. Formation and Stability of a Vinyl Carbanion at the Active Site of Orotidine 5‘-Monophosphate Decarboxylase: pKaof the C-6 Proton of Enzyme-Bound UMP. DOI:10.1021/ja710384t. PMID:18186641.
- Hu H et al. (2008), J Am Chem Soc, 130, 14493-14503. Mechanism of OMP Decarboxylation in Orotidine 5′-Monophosphate Decarboxylase. DOI:10.1021/ja801202j. PMID:18839943.
- Wepukhulu WO et al. (2008), Org Biomol Chem, 6, 4533-. A substantial oxygen isotope effect at O2 in the OMP decarboxylase reaction: Mechanistic implications. DOI:10.1039/b812979g. PMID:19039361.
- Toth K et al. (2007), J Am Chem Soc, 129, 12946-12947. Product Deuterium Isotope Effect for Orotidine 5‘-Monophosphate Decarboxylase: Evidence for the Existence of a Short-Lived Carbanion Intermediate. DOI:10.1021/ja076222f. PMID:17918849.
- Wu N et al. (2002), Biochemistry, 41, 4002-4011. Mapping the Active Site−Ligand Interactions of Orotidine 5‘-Monophosphate Decarboxylase by Crystallography†,‡. DOI:10.1021/bi015758p. PMID:11900543.
- Miller BG et al. (2002), Annu Rev Biochem, 71, 847-885. Catalytic Proficiency: The Unusual Case of OMP Decarboxylase. DOI:10.1146/annurev.biochem.71.110601.135446. PMID:12045113.
- Begley TP et al. (2000), Curr Opin Struct Biol, 10, 711-718. The structural basis for the remarkable catalytic proficiency of orotidine 5′-monophosphate decarboxylase. DOI:10.1016/s0959-440x(00)00148-2. PMID:11114509.
- Harris P et al. (2000), Biochemistry, 39, 4217-4224. Structural Basis for the Catalytic Mechanism of a Proficient Enzyme: Orotidine 5‘-Monophosphate Decarboxylase†,‡. DOI:10.1021/bi992952r. PMID:10757968.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys62A | hydrogen bond donor |
| Asp60A | hydrogen bond acceptor, repulsive charge-charge interaction, electrostatic destabiliser |
| Asp65B | hydrogen bond acceptor, electrostatic stabiliser |
| Arg215A | electrostatic destabiliser |
| Thr123A (main-N) | electrostatic stabiliser |
| Lys33A | electrostatic destabiliser |
| Arg215A | hydrogen bond donor |
| Lys62A | electrostatic stabiliser |
| Thr123A (main-N) | hydrogen bond donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, overall reactant used, overall product formed, decarboxylation, rate-determining stepCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys62A | hydrogen bond donor |
| Asp60A | hydrogen bond acceptor, activator |
| Asp65B | hydrogen bond acceptor, activator |
| Lys33A | electrostatic stabiliser, hydrogen bond donor |
| Lys62A | proton donor |
Chemical Components
native state of enzyme regenerated, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys62A | hydrogen bond acceptor, hydrogen bond donor |
| Asp60A | hydrogen bond acceptor, activator |
| Asp65B | hydrogen bond acceptor, activator |
| Lys33A | hydrogen bond donor, electrostatic stabiliser |
| Lys62A | proton acceptor |
Chemical Components
proton transfer, native state of enzyme regeneratedIntroduction
This is the carbene mechanism, lack of deuterium isotope effect and failure to detect the addition of a nucleophile has been used to suggest that this mechanism is not favoured. There is also no evidence of what residues could be acting as the proton donor or nucleophile in the other mechanistic suggestions. In the carbene mechanism, protonation of the O4 carbonyl generates a carbocation at C6 of the substrate. Decarboxylation would then generate a stabilised carbene, rather than the high-energy vinyl carbanion
Catalytic Residues Roles
| UniProt | PDB* (1dbt) | ||
| Lys62 | Lys62A | Acts as a general acid/base. Its pKa is modified through electrostatic interactions with the neighbouring carboxyl groups of Asp60 and Asp65(B). | proton acceptor, hydrogen bond donor, electrostatic stabiliser, proton donor |
| Arg215 | Arg215A | Binds the phosphate group, aids in stabilising the negatively charged transition state via long range interactions. | hydrogen bond donor, electrostatic destabiliser |
| Lys33 | Lys33A | Part of the catalytic tetrad. | electrostatic destabiliser |
| Asp60, Asp65 | Asp60A, Asp65B | The carboxylate side chains remain deprotonated and anionic within close proximity to the substrate carboxyl group. Although the groups repel one another, the strength of the surrounding hydrogen bonding network directs them towards one another. These repulsive interactions increase the energy of the groundstate so it resembles that of the transition state and therefore reduces the reaction activation barrier while also stabilising the polar transition state. | repulsive charge-charge interaction, hydrogen bond acceptor, electrostatic destabiliser |
| Thr123 (main-N) | Thr123A (main-N) | The backbone NH of Ser 127 (in M. thermoautotrophicus, Thr 123 here) increases as negative charge is delocalised to O4, this interaction could provide significant stabilisation of the intermediate. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
inferred reaction step, overall reactant used, proton transfer, decarboxylation, overall product formed, native state of enzyme regeneratedReferences
- Begley TP et al. (2000), Curr Opin Struct Biol, 10, 711-718. The structural basis for the remarkable catalytic proficiency of orotidine 5′-monophosphate decarboxylase. DOI:10.1016/s0959-440x(00)00148-2. PMID:11114509.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp65B | electrostatic stabiliser, hydrogen bond acceptor |
| Asp60A | electrostatic destabiliser, repulsive charge-charge interaction, hydrogen bond acceptor |
| Lys62A | hydrogen bond donor |
| Arg215A | electrostatic destabiliser |
| Thr123A (main-N) | electrostatic stabiliser |
| Lys33A | electrostatic destabiliser |
| Arg215A | hydrogen bond donor |
| Lys62A | electrostatic stabiliser |
| Thr123A (main-N) | hydrogen bond donor |
| Lys62A | proton donor |
Chemical Components
inferred reaction step, overall reactant used, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp65B | electrostatic stabiliser, hydrogen bond acceptor |
| Asp60A | electrostatic destabiliser, repulsive charge-charge interaction, hydrogen bond acceptor |
| Lys62A | hydrogen bond donor |
| Arg215A | electrostatic destabiliser |
| Thr123A (main-N) | electrostatic stabiliser |
| Lys33A | electrostatic destabiliser |
| Arg215A | hydrogen bond donor |
| Lys62A | electrostatic stabiliser |
| Thr123A (main-N) | hydrogen bond donor |
Chemical Components
inferred reaction step, decarboxylation, overall product formed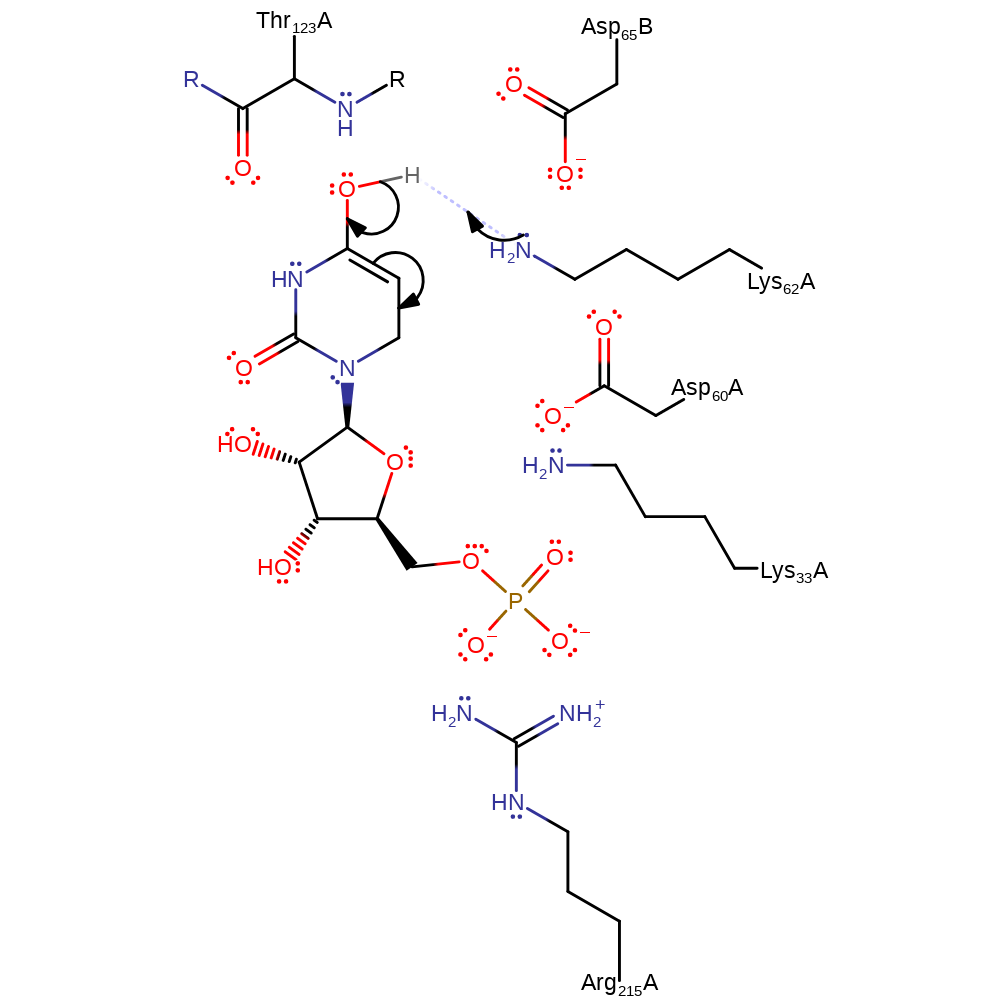
Step 3. Lys62 abstracts a proton from the intermediate to generate the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr123A (main-N) | hydrogen bond donor |
| Lys62A | electrostatic stabiliser |
| Arg215A | hydrogen bond donor |
| Lys33A | electrostatic destabiliser |
| Thr123A (main-N) | electrostatic stabiliser |
| Arg215A | electrostatic destabiliser |
| Lys62A | hydrogen bond donor |
| Asp60A | hydrogen bond acceptor, repulsive charge-charge interaction, electrostatic destabiliser |
| Asp65B | hydrogen bond acceptor, electrostatic stabiliser |
| Lys62A | proton acceptor |
Chemical Components
proton transfer, inferred reaction step, native state of enzyme regeneratedIntroduction
This is the nucleophilic addition mechanism, which due to lack of evidence of deuterium isotope effect and failure to detect the addition of a nucleophile has been suggested against. There is also no evidence of what residues could be acting as the nucleophile. In this mechanism the decarboxylation is proceeded by the initial addition of a nucleophile to C5 of OMP and the formation of the vinyl carbanion is avoided by the concerted elimination of carbon dioxide and the active site nucleophile.
Catalytic Residues Roles
| UniProt | PDB* (1dbt) | ||
| Lys62 | Lys62A | Acts as a general acid/base (cf primary mechanism proposal). Its pKa is modified through electrostatic interactions with the neighbouring carboxyl groups of Asp60 and Asp65(B). This residue has been selected as the nucleophile in this proposal based on its position relative to the substrate. | hydrogen bond donor, nucleophile, nucleofuge, activator, electrostatic stabiliser |
| Arg215 | Arg215A | Binds the phosphate group, aids in stabilising the negatively charged transition state via long range interactions. | hydrogen bond donor, electrostatic destabiliser |
| Lys33 | Lys33A | Part of the catalytic tetrad. | electrostatic destabiliser, proton acceptor, proton donor |
| Asp60, Asp65 | Asp60A, Asp65B | The carboxylate side chains remain deprotonated and anionic within close proximity to the substrate carboxyl group. Although the groups repel one another, the strength of the surrounding hydrogen bonding network directs them towards one another. These repulsive interactions increase the energy of the groundstate so it resembles that of the transition state and therefore reduces the reaction activation barrier while also stabilising the polar transition state. | repulsive charge-charge interaction, activator, hydrogen bond acceptor, electrostatic destabiliser |
| Thr123 (main-N) | Thr123A (main-N) | The backbone NH of Ser 127 (in M. thermoautotrophicus, Thr 123 here) increases as negative charge is delocalised to O4, this interaction could provide significant stabilisation of the intermediate. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, bimolecular nucleophilic addition, inferred reaction step, aromatic unimolecular elimination by the conjugate base, decarboxylation, overall product formed, native state of enzyme regeneratedReferences
- Begley TP et al. (2000), Curr Opin Struct Biol, 10, 711-718. The structural basis for the remarkable catalytic proficiency of orotidine 5′-monophosphate decarboxylase. DOI:10.1016/s0959-440x(00)00148-2. PMID:11114509.

Step 1. The enzyme initiates a nucleophilic attack on the substrate. The identity of the nucleophile is unknown and assumed to be Lys62 due to its proximity in the crystal structure.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr123A (main-N) | hydrogen bond donor |
| Lys62A | electrostatic stabiliser |
| Arg215A | hydrogen bond donor |
| Lys33A | electrostatic destabiliser |
| Thr123A (main-N) | electrostatic stabiliser |
| Arg215A | electrostatic destabiliser |
| Lys62A | hydrogen bond donor |
| Asp60A | hydrogen bond acceptor, repulsive charge-charge interaction, electrostatic destabiliser |
| Asp65B | hydrogen bond acceptor, electrostatic stabiliser |
| Lys62A | nucleophile |
| Lys33A | proton donor |
Chemical Components
proton transfer, overall reactant used, ingold: bimolecular nucleophilic addition, inferred reaction step
Step 2. Decarboxylation with concomitant elimination of the catalytic nucleophile.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr123A (main-N) | electrostatic stabiliser |
| Asp65B | electrostatic stabiliser |
| Asp60A | electrostatic destabiliser |
| Arg215A | electrostatic destabiliser |
| Lys33A | electrostatic destabiliser |
| Lys62A | nucleofuge |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, decarboxylation, overall product formed, inferred reaction step
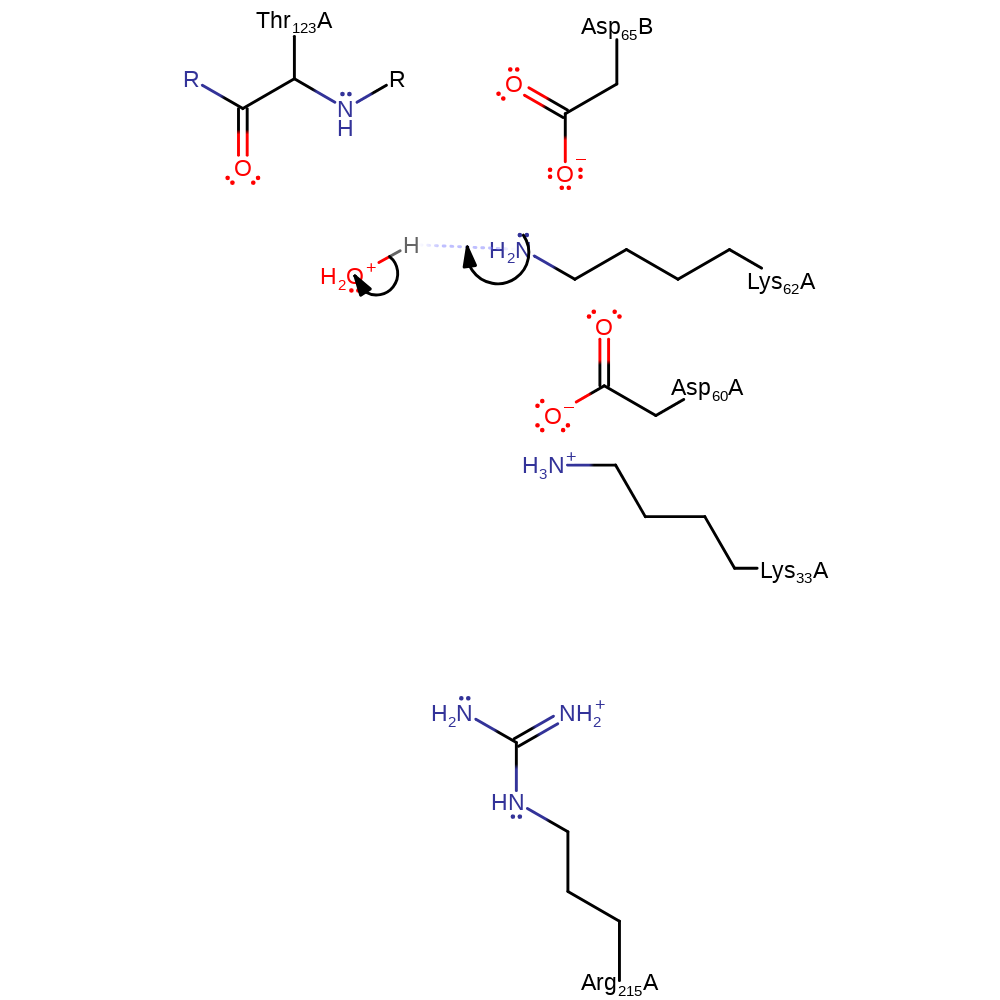
Step 3. Inferred return step to regenerate the protonation state of Lys33.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp60A | activator |
| Lys62A | activator |
| Asp65B | activator |
| Lys33A | proton acceptor |






 Download:
Download: 

 Download:
Download:  Download:
Download: