Histone acetyltransferase (MYST family)
ESA1 Histone acyl transferase is able to catalyse the acylation of histone proteins at specific lysine residues, using acetyl coA as the acyl donor for the reaction. Acylation of histone proteins is important in the regulatation of DNA transcription, as it causes the chromasones to condense and become inaccessible to the enzymes required for unwinding and unzipping the DNA. As a result study of the mechanism by which acylation occurs is of interest to geneticists, and opens the possibility of manipulation of gene expression by designing activators or inhibitors of the enzymes catalysing the process.
Histone acyl transferases in eukaryotes show much sequence homology, and can consequently be assigned to a superfamily (HAT). However, the mechanism of reaction has been previously thought to be not the same where the MYST subfamily, of which ESA1 is a representative, have an acylated enzyme intermediate in the reaction mechanism. Despite this, it has been proposed more recently ESA1 when in complex with two accessory proteins does in fact work proceed with direct attack from the target lysine residue on the acetyl-COA. It has been suggested the catalytic mechanism may reflect the enzyme's physiological surroundings.
Reference Protein and Structure
- Sequence
-
Q08649
 (2.3.1.48)
(2.3.1.48)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1mj9
- Crystal structure of yeast Esa1(C304S) mutant complexed with Coenzyme A
(2.5 Å)



- Catalytic CATH Domains
-
3.40.630.30
 (see all for 1mj9)
(see all for 1mj9)
Enzyme Reaction (EC:2.3.1.48)
Enzyme Mechanism
Introduction
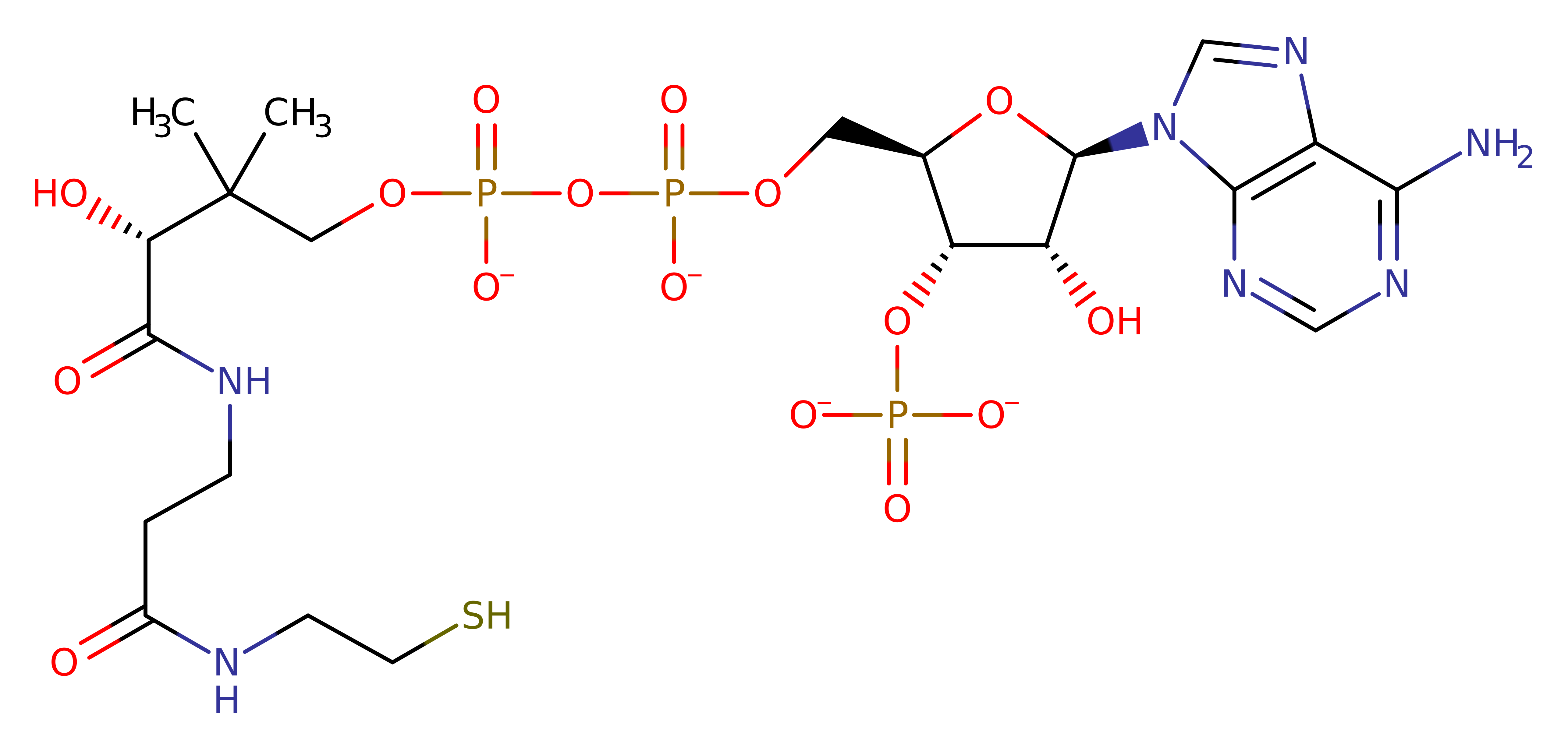
One reaction mechanism proposes it obeys ping-pong kinetics, with initial nucleophilic attack from Cys 304 on the acetyl-coA assisted by deprotonation of Cys 304 by the general base Glu 338. This results in the transfer of the acyl moiety to Cys 304. Histone binds and the target lysine is deprotonated by Glu 338 and then acts as a nucleophile, attacking the acyl moiety and resulting in Cys 304 acting as a leaving group. This transfers the acyl moiety to the Lysines epsilon NH group and forms the final product.
Catalytic Residues Roles
| UniProt | PDB* (1mj9) | ||
| Cys304 | Ser304(147)A | Acts as nucleophile to covalently attach to the acyl group of acetyl coA. Then acts as leaving group to allow transfer of the acyl group to the target lysine residue. | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor |
| Glu338 | Glu338(181)A | Acts as general acid base first in the deprotonation of Cys 304 to allow it to act as a nucleophile, then in the deprotonation of the target lysine to allow it to act as a nucleophile. | activator, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, enzyme-substrate complex formation, overall reactant used, intermediate formation, overall product formed, enzyme-substrate complex cleavage, intermediate terminated, inferred reaction step, native state of enzyme regeneratedReferences
- Yan Y et al. (2002), Nat Struct Biol, 9, 862-869. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. DOI:10.1038/nsb849. PMID:12368900.
- Cortopassi WA et al. (2016), J Mol Graph Model, 67, 69-84. Mechanisms of histone lysine-modifying enzymes: A computational perspective on the role of the protein environment. DOI:10.1016/j.jmgm.2016.04.011. PMID:27258188.
- Marmorstein R et al. (2014), Cold Spring Harb Perspect Biol, 6, a018762-. Writers and readers of histone acetylation: structure, mechanism, and inhibition. DOI:10.1101/cshperspect.a018762. PMID:24984779.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser304(147)A | proton donor |
| Glu338(181)A | proton acceptor |
Chemical Components
proton transfer
Step 2. Deprotonated Cys304 performs nucleophilic attack on the terminal acetyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser304(147)A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, enzyme-substrate complex formation, overall reactant used, intermediate formation
Step 3. Proton transfer from Glu338 to CoA, which then leaves the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser304(147)A | covalently attached |
| Glu338(181)A | proton donor |
Chemical Components
proton transfer, overall product formed
Step 4. Histone binds. Glu338 acts as a general base to deprotonate the histone lysine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu338(181)A | activator |
| Ser304(147)A | covalently attached |
| Glu338(181)A | proton acceptor |
Chemical Components
proton transfer, overall reactant used
Step 5. Histone lysine acts as a nucleophile to attack the enzyme-substrate intermediate, acetylating itself.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser304(147)A | proton acceptor, nucleofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution, enzyme-substrate complex cleavage, intermediate terminated, overall product formed, inferred reaction stepCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu338(181)A | proton donor |
Chemical Components
proton transfer, native state of enzyme regeneratedIntroduction
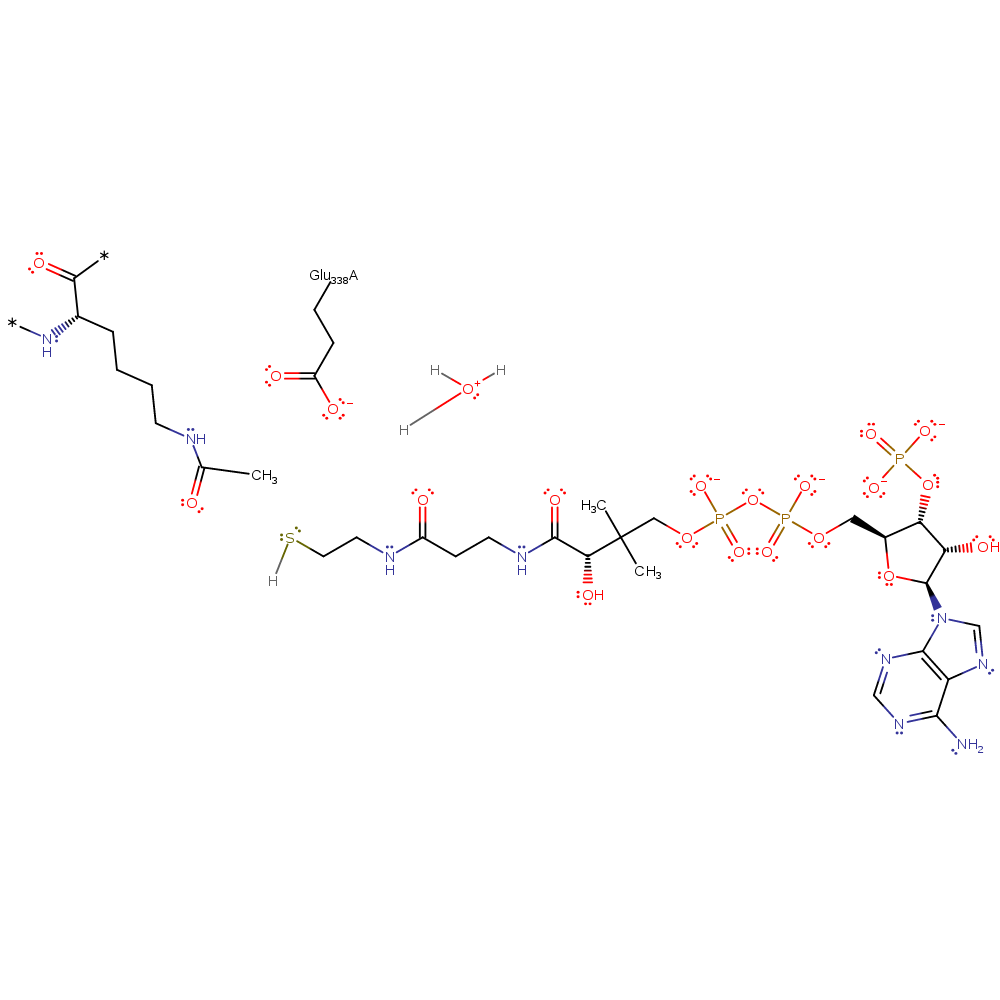
More recent mutagenesis studies have ruled out Cys304 as a catalytic residue in the Esa1 that physiologically associates to two accessory proteins to form the complex piccolo NuA4. Therefore, this mechanism proposal is different to the one seen in the other proposed mechanism. Esa1 is proposed to share a similar mechanism to Gcn5 histone acetyl transferase. Glu338 acts as a base catalyst to deprotonate the histone lysine's amine group. This promotes the direct nucleophilic attack of the lysine on the acetyl group on Acetyl-CoA to form a tetrahedral intermediate. This intermediate collapses to give the products of acetylated lysine and CoA.
Catalytic Residues Roles
| UniProt | PDB* (1mj9) | ||
| Glu338 | Glu338(181)A | Acts as a general base to promote the histone lysine nucleophilic attack on Acetyl-CoA. | activator, proton acceptor, proton donor |
Chemical Components
proton transfer, overall reactant used, bimolecular nucleophilic addition, intermediate formation, unimolecular elimination by the conjugate base, intermediate terminated, overall product formed, native state of enzyme regenerated, inferred reaction stepReferences
- Berndsen CE et al. (2007), Biochemistry, 46, 623-629. Catalytic mechanism of a MYST family histone acetyltransferase. DOI:10.1021/bi602513x. PMID:17223684.
- Cortopassi WA et al. (2016), J Mol Graph Model, 67, 69-84. Mechanisms of histone lysine-modifying enzymes: A computational perspective on the role of the protein environment. DOI:10.1016/j.jmgm.2016.04.011. PMID:27258188.
- Marmorstein R et al. (2014), Cold Spring Harb Perspect Biol, 6, a018762-. Writers and readers of histone acetylation: structure, mechanism, and inhibition. DOI:10.1101/cshperspect.a018762. PMID:24984779.

Step 1. Glu338 deprotonates histone lysine amine group, promoting its nucleophilicity.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu338(181)A | activator, proton acceptor |
Chemical Components
proton transfer, overall reactant used
Step 2. Lysine performs a nucleophilic attack on the acetyl group on acetyl-CoA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation
Step 3. Tetrahedral intermediate collapses to form the products CoA and acetylated lysine on the histone protein.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate terminated, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu338(181)A | proton donor |






 Download:
Download: 
 Download:
Download: