Malate dehydrogenase (type 2)
Malate dehydrogenase catalysis the readily reversible dehydrogenation of malate to oxaloacetate. As the activated cofactor/substrate complex has a net negative charge, a positive counter-charge is provided by conserved arginines in the active site. By this mechanism, malate dehydrogenase uses charge balancing to achieve fivefold orders of magnitude in discrimination between potential substrates.
Reference Protein and Structure
- Sequence
-
P11708
 (1.1.1.37)
(1.1.1.37)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Sus scrofa (pig)

- PDB
-
4mdh
- REFINED CRYSTAL STRUCTURE OF CYTOPLASMIC MALATE DEHYDROGENASE AT 2.5-ANGSTROMS RESOLUTION
(2.5 Å)



- Catalytic CATH Domains
-
3.90.110.10
 (see all for 4mdh)
(see all for 4mdh)
Enzyme Reaction (EC:1.1.1.37)
Enzyme Mechanism
Introduction
The enzyme uses a catalytic Asp-His dyad. The C2 position of malate is situated above the C4 position of the nicotinamide moiety of NAD+. His186 is in position to abstract a proton from the hydroxyl group attached to C2 which then forms a carbonyl group with the transfer of a hydride to NAD+. Thus oxaloacetate and NADH are formed and may leave the active site. Asp158 hydrogen bonds to His186 stabilising the protonated imidazole ring.
Catalytic Residues Roles
| UniProt | PDB* (4mdh) | ||
| His187 | His186(187)A | Abstracts a proton from the hydroxyl group at C2. | proton acceptor, proton donor |
| Asp159 | Asp158(159)A | Electrostatic stabilisation of the charge formed on His186 during the dehydrogenation. | electrostatic stabiliser |
Chemical Components
proton transfer, hydride transfer, overall product formed, overall reactant used, aromatic bimolecular nucleophilic addition, inferred reaction step, native state of enzyme regeneratedReferences
- Birktoft JJ et al. (1989), Biochemistry, 28, 6065-6081. Refined crystal structure of cytoplasmic malate dehydrogenase at 2.5-.ANG. resolution. DOI:10.1021/bi00440a051. PMID:2775751.
- Chapman AD et al. (1999), J Mol Biol, 285, 703-712. Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, α-Ketomalonate and TetrahydoNAD. DOI:10.1006/jmbi.1998.2357. PMID:10075524.
- Lemaire M et al. (1996), Eur J Biochem, 236, 947-952. The Catalytic Site of Chloroplastic NADP-Dependent Malate Dehydrogenase Contains A His/Asp Pair. DOI:10.1111/j.1432-1033.1996.00947.x. PMID:8665917.
- Goward CR et al. (1994), Protein Sci, 3, 1883-1888. Malate dehydrogenase: A model for structure, evolution, and catalysis. DOI:10.1002/pro.5560031027. PMID:7849603.
- Birktoft JJ et al. (1983), J Biol Chem, 258, 472-482. The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. DOI:10.2210/pdb2mdh/pdb. PMID:6848515.

Step 1. His186 abstracts a proton from the hydroxyl group of L-malate. A hydride is transferred from L-malate to NAD+ in an aromatic bimolecular nucleophilic addition reaction, forming NADH and the product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp158(159)A | electrostatic stabiliser |
| His186(187)A | proton acceptor |
Chemical Components
proton transfer, hydride transfer, overall product formed, overall reactant used, ingold: aromatic bimolecular nucleophilic addition
Step 2. In an inferred reaction step His186 is deprotonated to regenerate the native state of the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His186(187)A | proton donor |
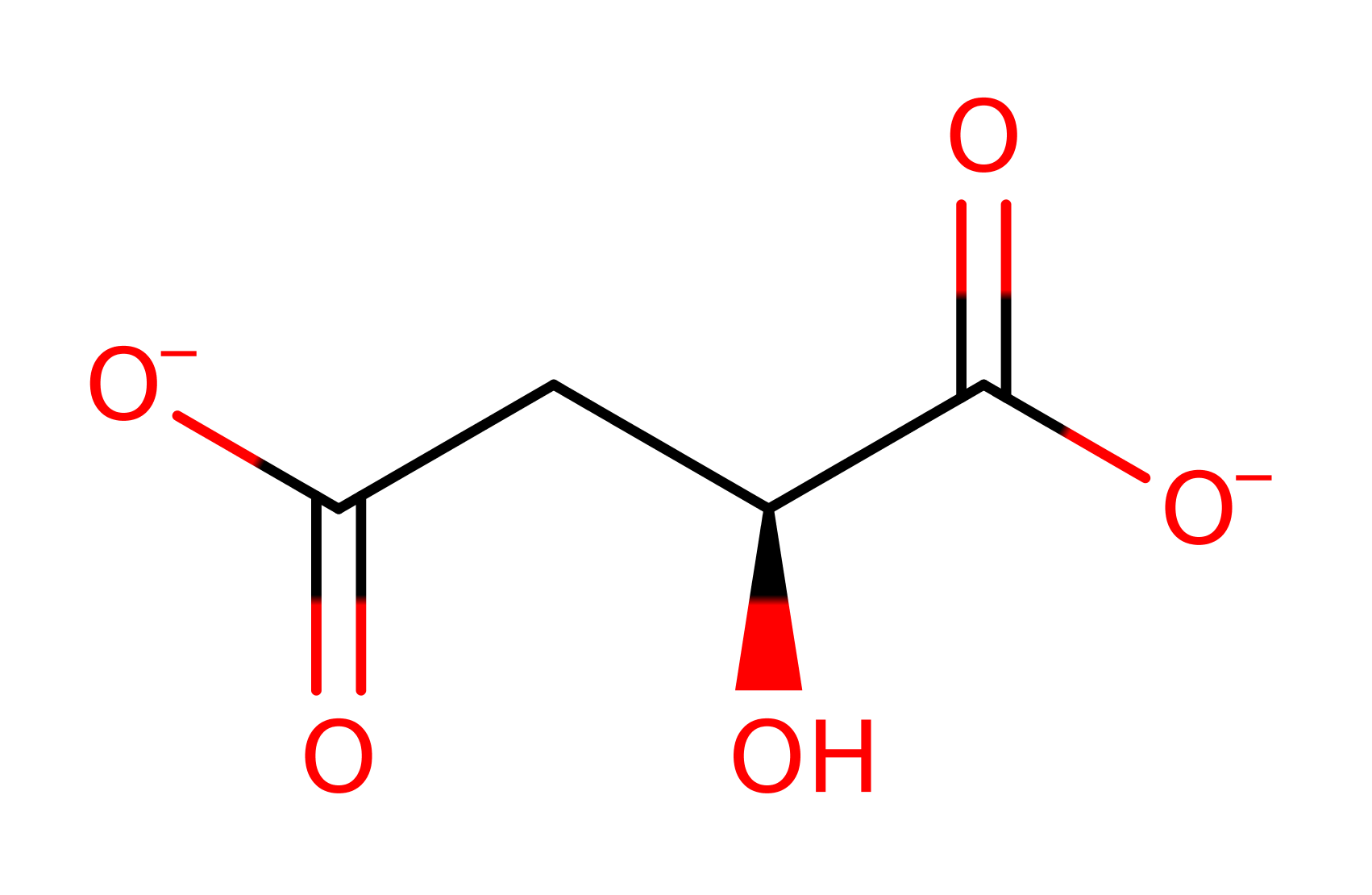




 Download:
Download: