G-protein alpha subunit, group I (GTPase)
The alpha subunits of heterotrimeric G proteins are members of the Ras superfamily of GTP hydrolases and function in signal transduction. They are activated by replacement of bound GDP by GTP, a process promoted by activated 7-transmembrane receptors, and by subsequent dissociation of the alpha subunit from the beta and gamma subunits. The G-alpha subunit remains active until its GTPase activity converts the bound GTP back to GDP.
The alpha subunit of the G protein Gi1 is expressed in many different tissues and is activated by the alpha-2 adrenergic and M-2 muscarinic cholinergic receptors, amongst others. Its downstream effectors include particular isoforms of adenylate cyclase; types V and VI are particularly sensitive to inhibition by G-alpha-i1.
The mechanism of enzymatic GTP hydrolysis (and similar phosphoryl transfer reactions) has been subject to significant debate and still remains to be firmly established. More recent models have proposed a dissociative-like process, in which the transition state shows significant cleavage of the gamma-beta phosphoanhydride bond and relatively little bond formation between the gamma phosphate and the nucleophilic water molecule. (See, for example, the reference pubmed ID 15236956 for a recent discussion).
Reference Protein and Structure
- Sequence
-
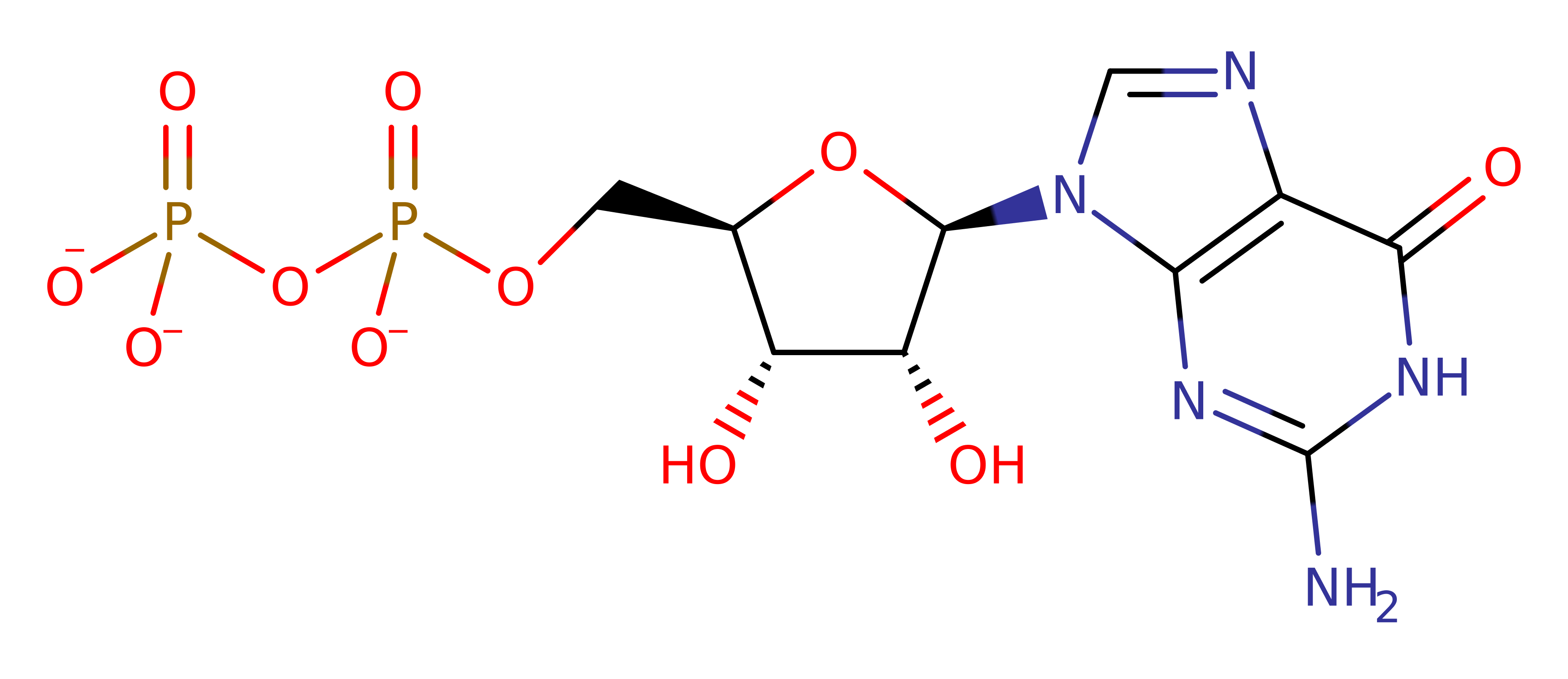
P10824

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rattus norvegicus (Norway rat)

- PDB
-
1bh2
- A326S MUTANT OF AN INHIBITORY ALPHA SUBUNIT
(2.1 Å)



- Catalytic CATH Domains
-
1.10.400.10
 3.40.50.300
3.40.50.300  (see all for 1bh2)
(see all for 1bh2)
- Cofactors
- Magnesium(2+) (1)
Enzyme Mechanism
Introduction
Assuming a dissociative mechanism for G-alpha-i1, the key catalytic residue is Arg 278 (side chain) which stabilise accumulation of negative charge on the beta-gamma bridging oxygen as it departs from the gamma phosphate in the transition state. In addition the binding of RGS causes Arg 278 to shift from a monodenate coordination to a bidentate coordination with both the gamma and alpha phosphate. This withdraws electrons from the gamma phosphate to the beta phosphate and also results in the gamma phosphate being more planar. This makes the gamma phosphate more alike to the transition state and as a result will favour a dissociative mechanism and so the bond between the gamma and beta phosphate will dissociate. An Mg2+ ion may play a similar role in stabilising accumulation of negative charge on the beta phosphate, although whether such metal ions are catalytic has been disputed (see for example the reference PMID:9383480). Gln 204 has a role in polarising and orientating the lytic water molecule, although in a dissociative mechanism activation of the water molecule would not make a large contribution to catalysis (see the reference PMID:8710841). There is some evidence that conformational changes immediately prior to hydrolysis are required to properly position the catalytic residues.
Catalytic Residues Roles
| UniProt | PDB* (1bh2) | ||
| Thr48 | Thr48(17)A | Stabilises the alpha phosphate and the twist in conformation between once Arg 178 has coordinated. | electrostatic stabiliser |
| Asp200 | Asp200(169)A | Positions Magnesium coordinated water | electrostatic stabiliser |
| Glu43 | Glu43(12)A | Stabilises Arg 178 by forming a salt bridge | electrostatic stabiliser |
| Arg178 | Arg178(147)A | Proposed to stabilise accumulation of negative charge on the beta-gamma bridging oxygen as this atom departs from the gamma phosphate in the transition state and coordinate to the the alpha and gamma phosphates. Which results in the withdrawing of electronic charge from the gamma phosphate and it adopting a planar geometry which results in the favouring of a dissociative mechanism. | electrostatic stabiliser |
| Gln204 | Gln204(173)A | Proposed to position and polarise the lytic water molecule. | electrostatic stabiliser |
Chemical Components
proton transfer, elimination (not covered by the Ingold mechanisms), heterolysis, intermediate formation, overall reactant used, bimolecular nucleophilic addition, intermediate collapse, overall product formed, inferred reaction stepReferences
- Coleman DE et al. (1994), Science, 265, 1405-1412. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. DOI:10.1126/science.8073283. PMID:8073283.
- Gerwert K et al. (2017), Biol Chem, 398, 523-533. Common mechanisms of catalysis in small and heterotrimeric GTPases and their respective GAPs. DOI:10.1515/hsz-2016-0314. PMID:28245182.
- Mann D et al. (2016), Proc Natl Acad Sci U S A, 113, E8041-E8050. Mechanism of the intrinsic arginine finger in heterotrimeric G proteins. DOI:10.1073/pnas.1612394113. PMID:27911799.
- Knihtila R et al. (2015), J Biol Chem, 290, 31025-31036. Neutron Crystal Structure of RAS GTPase Puts in Question the Protonation State of the GTP γ-Phosphate. DOI:10.1074/jbc.M115.679860. PMID:26515069.
- Li G et al. (2004), J Mol Biol, 340, 921-932. GTP Hydrolysis Mechanism of Ras-like GTPases. DOI:10.1016/j.jmb.2004.06.007. PMID:15236956.
- Thomas CJ et al. (2004), Proc Natl Acad Sci U S A, 101, 7560-7565. Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein -subunit. DOI:10.1073/pnas.0304091101. PMID:15128951.
- Coleman DE et al. (1999), Methods Enzymol, 308, 70-92. [4] Reaction dynamics of G-protein catalyzed hydrolysis of GTP as viewed by X-ray crystallographic snapshots of Giα1. DOI:10.1016/s0076-6879(99)08006-4. PMID:10507001.
- Maegley KA et al. (1996), Proc Natl Acad Sci U S A, 93, 8160-8166. Ras-catalyzed hydrolysis of GTP: a new perspective from model studies. DOI:10.1073/pnas.93.16.8160. PMID:8710841.
- Admiraal SJ et al. (1995), Chem Biol, 2, 729-739. Mapping the transition state for ATP hydrolysis: implications for enzymatic catalysis. DOI:10.1016/1074-5521(95)90101-9. PMID:9383480.

Step 1. The substrate accepts a proton and and the bidentate coordination of Arg178 shifts the charge away from the gamma phosphate resulting in the breaking of the phosphodiester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln204(173)A | electrostatic stabiliser |
| Arg178(147)A | electrostatic stabiliser |
| Thr48(17)A | electrostatic stabiliser |
| Glu43(12)A | electrostatic stabiliser |
| Asp200(169)A | electrostatic stabiliser |
Chemical Components
proton transfer, elimination (not covered by the Ingold mechanisms), heterolysis, intermediate formation, overall reactant used
Step 2. The hydroxide formed nucleophilically attacks the gamma phosphate. The beta phosphate most likely loses a proton to the surrounding solution to produce the favourable protonation state of the product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu43(12)A | electrostatic stabiliser |
| Thr48(17)A | electrostatic stabiliser |
| Arg178(147)A | electrostatic stabiliser |
| Asp200(169)A | electrostatic stabiliser |
| Gln204(173)A | electrostatic stabiliser |




 Download:
Download: