Myosin ATPase
Myosin is the key enzyme in transducing the energy from ATP hydrolysis into directed movement. Muscle contraction involves the relative movements of myosin and actin filaments. This movement is achieved by the interaction of the globular heads of myosin with actin, and is driven by the hydrolysis of ATP in the myosin head domains.
Reference Protein and Structure
- Sequence
-
P08799

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Dictyostelium discoideum (Slime mould)

- PDB
-
1vom
- COMPLEX BETWEEN DICTYOSTELIUM MYOSIN AND MGADP AND VANADATE AT 1.9A RESOLUTION
(1.9 Å)



- Catalytic CATH Domains
-
3.40.850.10
 1.20.58.530
1.20.58.530  (see all for 1vom)
(see all for 1vom)
- Cofactors
- Magnesium(2+) (1)
Enzyme Mechanism
- Mechanism Proposal 1 ☆☆
- Mechanism Proposal 2 ☆☆
- Mechanism Proposal 3 ☆☆
- Mechanism Proposal 4 ☆☆
- Mechanism Proposal 5 ☆☆
- Mechanism Proposal 6 ☆☆
Introduction
Binding of ATP to the active site induces cleft closure with Glu 459 moving to form a salt bridge with Arg 238. The 'lytic' water molecule is released from Arg 238 and positioned for nucleophilic attack on the gamma phosphate by interactions with Ser 237 and with a second water molecule that interacts wih Glu 459 and the backbone oxygen of Gly 457. Departure of the gamma phosphate of ATP from the beta phosphate is promoted by a hydrogen bond from a gamma phosphate oxygen to the backbone NH group of Gly 457; this hydrogen bond forms specifically in the transition state. Attack by the lytic water molecule on the gamma phosphate is promoted by deprotonation of this water by the second water (which ends up as a hydronium ion, one of the reaction products as determined experimentally). Positive charge accumulation on this second water molecule is stabilised by its interactions with Glu 459 and with Gly 457. Accumulation of negative charge on the beta-gamma bridging oxygen as it departs from the gamma phosphate is stabilised by interactions of this atom to the side chain NH2 of Asn 233 and the backbone NH of Gly 182.
Catalytic Residues Roles
| UniProt | PDB* (1vom) | ||
| Asn233 | Asn233A | Side chain amide stabilises accumulation of negative charge on the beta-gamma bridging oxygen as this atom departs from the gamma phosphate. | electrostatic stabiliser |
| Gly182 (main-N) | Gly182A (main-N) | Backbone amide stabilises accumulation of negative charge on the beta-gamma bridging oxygen as this atom departs from the gamma phosphate. | electrostatic stabiliser |
| Gly457 (main-N) | Gly457A (main-N) | Backbone NH is proposed to form a hydrogen bond to a gamma phosphate oxygen specifically in the transition state. Backbone O is proposed to interact with and stabilise accumulation of positive charge on the water molecule that deprotonates the lytic water. | electrostatic stabiliser |
| Glu459 | Glu459A | Proposed to stabilise accumulation of positive charge on the water molecule that deprotonates the lytic water. | electrostatic stabiliser |
Chemical Components
References
- Onishi H et al. (2004), Biochemistry, 43, 3757-3763. On the Myosin Catalysis of ATP Hydrolysis†. DOI:10.1021/bi040002m. PMID:15049682.
- Onishi H et al. (2002), Proc Natl Acad Sci U S A, 99, 15339-15344. Nonlinear partial differential equations and applications: Early stages of energy transduction by myosin: Roles of Arg in Switch I, of Glu in Switch II, and of the salt-bridge between them. DOI:10.1073/pnas.242604099. PMID:12429851.
- Onishi H et al. (1997), Biochemistry, 36, 3767-3772. Functional Transitions in Myosin: Role of Highly Conserved Gly and Glu Residues in the Active Site†. DOI:10.1021/bi9630772. PMID:9092805.
- Smith CA et al. (1996), Biochemistry, 35, 5404-5417. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. DOI:10.1021/bi952633+. PMID:8611530.
- Fisher AJ et al. (1995), Biochemistry, 34, 8960-8972. X-ray Structures of the Myosin Motor Domain of Dictyostelium discoideum Complexed with MgADP.cntdot.BeFx and MgADP.cntdot.AlF4-. DOI:10.1021/bi00028a004. PMID:7619795.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu459A | electrostatic stabiliser |
| Gly182A (main-N) | electrostatic stabiliser |
| Gly457A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
Chemical Components
Introduction
A salt bridge is formed between Glu 459 and Arg 238 due to the binding of ATP to the active site which induces cleft closure with Glu 459. The 'lytic' water molecule is released from Arg 238 and positioned for nucleophilic attack on the gamma phosphate by interactions with Ser 237. For this mechanism water directly attacks the gamma phosphate of ATP. As ATP will deprotonate the water molecule itself to activate it so it can nucleophilically attack the phosphoryl. The negative charge that accumulates on the leaving group is stabilised by coordination to Mg2+ and hydrogen bonding to backbone amides of Gly182 and the side chain amide of Asn233.
Catalytic Residues Roles
| UniProt | PDB* (1vom) | ||
| Ser237, Thr186 | Ser237A, Thr186A | Forms Mg2+ binding site | metal ligand |
| Asn233, Gly182 (main-N) | Asn233A, Gly182A (main-N) | Stabilises the negative charge that accumulates on the leaving group by hydrogen bonding from their respective amide groups. | electrostatic stabiliser |
| Gly457 (main-N) | Gly457A (main-N) | Backbone NH is proposed to form a hydrogen bond to a gamma phosphate oxygen specifically in the transition state. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedReferences
- Kiani FA et al. (2015), Curr Opin Struct Biol, 31, 115-123. Advances in quantum simulations of ATPase catalysis in the myosin motor. DOI:10.1016/j.sbi.2015.04.006. PMID:26005996.
- Grigorenko BL et al. (2011), J Mol Graph Model, 31, 1-4. Minimum energy reaction profiles for ATP hydrolysis in myosin. DOI:10.1016/j.jmgm.2011.07.005. PMID:21839658.
- Schwarzl SM et al. (2006), Biochemistry, 45, 5830-5847. Insights into the chemomechanical coupling of the myosin motor from simulation of its ATP hydrolysis mechanism. DOI:10.1021/bi052433q. PMID:16669626.
- Li G et al. (2004), J Phys Chem B, 108, 3342-3357. Mechanochemical Coupling in Myosin: A Theoretical Analysis with Molecular Dynamics and Combined QM/MM Reaction Path Calculations. DOI:10.1021/jp0371783.
- Onishi H et al. (2004), Biochemistry, 43, 3757-3763. On the Myosin Catalysis of ATP Hydrolysis†. DOI:10.1021/bi040002m. PMID:15049682.
- Okimoto N et al. (2001), Biophys J, 81, 2786-2794. Theoretical Studies of the ATP Hydrolysis Mechanism of Myosin. DOI:10.1016/S0006-3495(01)75921-8.
- Bauer CB et al. (2000), J Biol Chem, 275, 38494-38499. X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain. DOI:10.1074/jbc.M005585200. PMID:10954715.

Step 1. The gamma phosphate directly deprotonates a water which activates it to attack the phopshate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly182A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly457A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 2. The pentacoordinate transition state collapse resulting in the cleavage of the phopshodiester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly182A (main-N) | electrostatic stabiliser |
| Gly457A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
| Asn233A | electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedIntroduction
Binding of ATP to the active site induces cleft closure with Glu 459 moving to form a salt bridge with Arg 238. The 'lytic' water molecule is released from Arg 238 and positioned for nucleophilic attack on the gamma phosphate by interactions with Ser 237 and with a second water molecule that interacts with Glu 459 and the backbone oxygen of Gly 457. The second water molecule protonates the gamma phosphate which then enables it to accept a proton from the "lytic water" which activates that water to nucleophilically attack the gamma phosphate. Which will produce the transition state which has a pentavalent gamma-phosphate in a bipyramidal configuration. The transition state collapses, cleaving the phosphodiester bond and the negative charge that accumulate on the leaving group is stabilised by the amide group of Gly 182 and Asn 233 and also by coordination to Mg2+ ion.
Catalytic Residues Roles
| UniProt | PDB* (1vom) | ||
| Ser237, Thr186 | Ser237A, Thr186A | Form Mg2+ binding site | metal ligand |
| Asn233, Gly182 (main-N) | Asn233A, Gly182A (main-N) | Stabilises the accumulated negative charge on the leaving group | electrostatic stabiliser |
| Gly457 (main-N) | Gly457A (main-N) | Backbone NH is proposed to form a hydrogen bond to a gamma phosphate oxygen specifically in the transition state. Backbone O is proposed to interact with and stabilise accumulation of positive charge on the water molecule that deprotonates the lytic water. | electrostatic stabiliser |
| Glu459 | Glu459A | Stabilises and positions the water that deprotonates the lytic water |
Chemical Components
overall reactant used, intermediate formation, bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedReferences
- Kiani FA et al. (2015), Curr Opin Struct Biol, 31, 115-123. Advances in quantum simulations of ATPase catalysis in the myosin motor. DOI:10.1016/j.sbi.2015.04.006. PMID:26005996.
- Kiani FA et al. (2014), Proc Natl Acad Sci U S A, 111, E2947-E2956. Catalytic strategy used by the myosin motor to hydrolyze ATP. DOI:10.1073/pnas.1401862111. PMID:25006262.
- Onishi H et al. (2004), Biochemistry, 43, 3757-3763. On the Myosin Catalysis of ATP Hydrolysis†. DOI:10.1021/bi040002m. PMID:15049682.

Step 1. The gamma phosphate directly deprotonates a water which activates it to deprotonate another water which can then nucleophilically attack the gamma phosphate
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly457A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly182A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
Chemical Components
overall reactant used, intermediate formation, ingold: bimolecular nucleophilic addition, proton transfer
Step 2. The pentavalent transition state collapses resulting in the cleavage of the phosphodiester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly182A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly457A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedIntroduction
The lytic water, which is released due to the formation of a salt bridge between Glu 459 and Arg 238 when ATP binds, can nucleophilically attack the gamma phosphate after it is deprotonated by Ser 236 which is deprotonated by the gamma phosphate. The pentavalent transition state formed will then collapse resulting in the cleavage of the phosphodiester bond. The negative charge that then accumulates on the leaving group is stabilised by the backbone and side chain amide groups on Gly 182 and Asn 233 and also the Mg2+ ion.
Catalytic Residues Roles
| UniProt | PDB* (1vom) | ||
| Ser236 | Ser236A | Acts as a general acid/base as is deprotonated by the gamma phosphate of ATP to enable it to accept a proton from a water which activates the water for nucleophilic attack. | proton relay, proton acceptor, proton donor |
| Ser237, Thr186 | Ser237A, Thr186A | Forms Mg2+ binding site | metal ligand |
| Asn233, Gly182 (main-N) | Asn233A, Gly182A (main-N) | Stabilises the accumulated negative charge on the leaving group | electrostatic stabiliser |
| Gly457 (main-N) | Gly457A (main-N) | Backbone NH is proposed to form a hydrogen bond to a gamma phosphate oxygen specifically in the transition state. | electrostatic stabiliser |
Chemical Components
overall reactant used, intermediate formation, bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedReferences
- Kiani FA et al. (2015), Curr Opin Struct Biol, 31, 115-123. Advances in quantum simulations of ATPase catalysis in the myosin motor. DOI:10.1016/j.sbi.2015.04.006. PMID:26005996.
- Yang Y et al. (2008), J Mol Biol, 381, 1407-1420. Extensive conformational transitions are required to turn on ATP hydrolysis in myosin. DOI:10.1016/j.jmb.2008.06.071. PMID:18619975.
- Schwarzl SM et al. (2006), Biochemistry, 45, 5830-5847. Insights into the chemomechanical coupling of the myosin motor from simulation of its ATP hydrolysis mechanism. DOI:10.1021/bi052433q. PMID:16669626.
- Li G et al. (2004), J Phys Chem B, 108, 3342-3357. Mechanochemical Coupling in Myosin: A Theoretical Analysis with Molecular Dynamics and Combined QM/MM Reaction Path Calculations. DOI:10.1021/jp0371783.
- Onishi H et al. (2004), Biochemistry, 43, 3757-3763. On the Myosin Catalysis of ATP Hydrolysis†. DOI:10.1021/bi040002m. PMID:15049682.

Step 1. The gamma phosphate deprotonates Ser236 which enables it to accept a proton from the lytic water which can now nucleophilically attack the the gamma phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly457A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly182A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
| Ser236A | proton acceptor, proton donor, proton relay |
Chemical Components
overall reactant used, intermediate formation, ingold: bimolecular nucleophilic addition, proton transfer
Step 2. The pentavalent transition state collapses resulting in the cleavage of the phosphodiester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly182A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly457A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedIntroduction
As ATP binds and induces cleft closure so that the lytic water can be released from Arg 238 as it forms a salt bridge with Glu 459; Ser 181 is deprotonated by the gamma phosphate. This enables it to accept a proton from the lytic water so that the water can then nucleophilically attack the gamma phosphate. The transition state will collapse resulting in the cleavage of the phosphodiester bond and the leaving group, beta-phosphate will be stabilised by the backbone amide of Gly 182 and the sidechain amide of Asn 233, as well as the magnesium ion.
Catalytic Residues Roles
| UniProt | PDB* (1vom) | ||
| Ser181 | Ser181A | Acts as a general acid/ base as is deprotonated by the gamma phosphate which enables it to activate the lytic water by deprotonating it so that the water can act as a nucleophile. | proton relay, proton acceptor, proton donor |
| Ser237, Thr186 | Ser237A, Thr186A | Forms the Mg2+ binding site | metal ligand |
| Asn233, Gly182 (main-N) | Asn233A, Gly182A (main-N) | Stabilises the accumulate negative charge on the leaving group, beta phosphate. | electrostatic stabiliser |
| Gly457 (main-N) | Gly457A (main-N) | Backbone NH is proposed to form a hydrogen bond to a gamma phosphate oxygen specifically in the transition state. | electrostatic stabiliser |
Chemical Components
overall reactant used, intermediate formation, bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedReferences
- Kiani FA et al. (2015), Curr Opin Struct Biol, 31, 115-123. Advances in quantum simulations of ATPase catalysis in the myosin motor. DOI:10.1016/j.sbi.2015.04.006. PMID:26005996.
- Grigorenko BL et al. (2011), J Mol Graph Model, 31, 1-4. Minimum energy reaction profiles for ATP hydrolysis in myosin. DOI:10.1016/j.jmgm.2011.07.005. PMID:21839658.
- Schwarzl SM et al. (2006), Biochemistry, 45, 5830-5847. Insights into the chemomechanical coupling of the myosin motor from simulation of its ATP hydrolysis mechanism. DOI:10.1021/bi052433q. PMID:16669626.
- Onishi H et al. (2004), Biochemistry, 43, 3757-3763. On the Myosin Catalysis of ATP Hydrolysis†. DOI:10.1021/bi040002m. PMID:15049682.

Step 1. Ser181 is deprotonated by the gamma phosphate which enables it to accept a proton from the lytic water. This activates the water to nucleophilically attack the gamma phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly457A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly182A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
| Ser181A | proton acceptor, proton donor, proton relay |
Chemical Components
overall reactant used, intermediate formation, ingold: bimolecular nucleophilic addition, proton transfer
Step 2. The pentavalent transition state collapses resulting in the cleavage of the phosphodiester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly182A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly457A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base, inferred reaction step, intermediate collapse, overall product formedIntroduction
Glu 459 can accept a proton from a water which enables it to accept a proton from the lytic water which activates that water to nucleophilially attack the gamma phosphate to produce the pentavalent transition state. Ser 181 then is deprotonated by the gamma phosphate which then enables it to accept a proton from a thrid water which will then accept a proton from Glu 459 and thus returning Glu 459 to its original protonation state. The protonation of the gamma phosphate also induces an elimination and as a result the phosphodiester bond is cleaved. The beta phosphate leaving group accumulates a negative charge which is stabilised by the backbone amide of Gly 182 and the side chain amide of Asn 233 as well as the magnesium ion.
Catalytic Residues Roles
| UniProt | PDB* (1vom) | ||
| Ser181 | Ser181A | protonates the gamma phosphate so that it can accept a proton from a water which enables said water to regenerate the original protonation state of Glu 459. | proton relay, proton acceptor, proton donor |
| Ser237, Thr186 | Ser237A, Thr186A | Forms Mg2+ binding site | metal ligand |
| Asn233, Gly182 (main-N) | Asn233A, Gly182A (main-N) | Stabilises the accumulated negative charge on the beta phosphate leaving group | electrostatic stabiliser |
| Gly457 (main-N) | Gly457A (main-N) | Backbone NH is proposed to form a hydrogen bond to a gamma phosphate oxygen specifically in the transition state. | electrostatic stabiliser |
| Glu459 | Glu459A | Acts as a general base as deprotonates a water which will deprotonate the lytic water so that it can nucleophilically attack the gamma phosphate. | proton acceptor, proton donor |
Chemical Components
overall reactant used, intermediate formation, bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate base, intermediate collapse, overall product formedReferences
- Kiani FA et al. (2015), Curr Opin Struct Biol, 31, 115-123. Advances in quantum simulations of ATPase catalysis in the myosin motor. DOI:10.1016/j.sbi.2015.04.006. PMID:26005996.
- Kiani FA et al. (2014), Proc Natl Acad Sci U S A, 111, E2947-E2956. Catalytic strategy used by the myosin motor to hydrolyze ATP. DOI:10.1073/pnas.1401862111. PMID:25006262.
- Onishi H et al. (2004), Biochemistry, 43, 3757-3763. On the Myosin Catalysis of ATP Hydrolysis†. DOI:10.1021/bi040002m. PMID:15049682.

Step 1. Glu 459 deprotonates a water which then enables this water to accept a proton from the lytic water. The deprotonated lytic water can then nucleophilically attack the gamma phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly457A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly182A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
| Glu459A | proton acceptor |
Chemical Components
overall reactant used, intermediate formation, ingold: bimolecular nucleophilic addition, proton transfer
Step 2. The gamma phosphate accepts a proton from Ser 181 which enables the regeneration of the native protonation state of Glu459 via a third water. Also the transition state collapse on protonation and results in the cleavage of the phosphodiester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly182A (main-N) | electrostatic stabiliser |
| Asn233A | electrostatic stabiliser |
| Gly457A (main-N) | electrostatic stabiliser |
| Ser237A | metal ligand |
| Thr186A | metal ligand |
| Ser181A | proton acceptor |
| Glu459A | proton donor |
| Ser181A | proton donor, proton relay |






 Download:
Download:  Download:
Download:  Download:
Download:  Download:
Download: 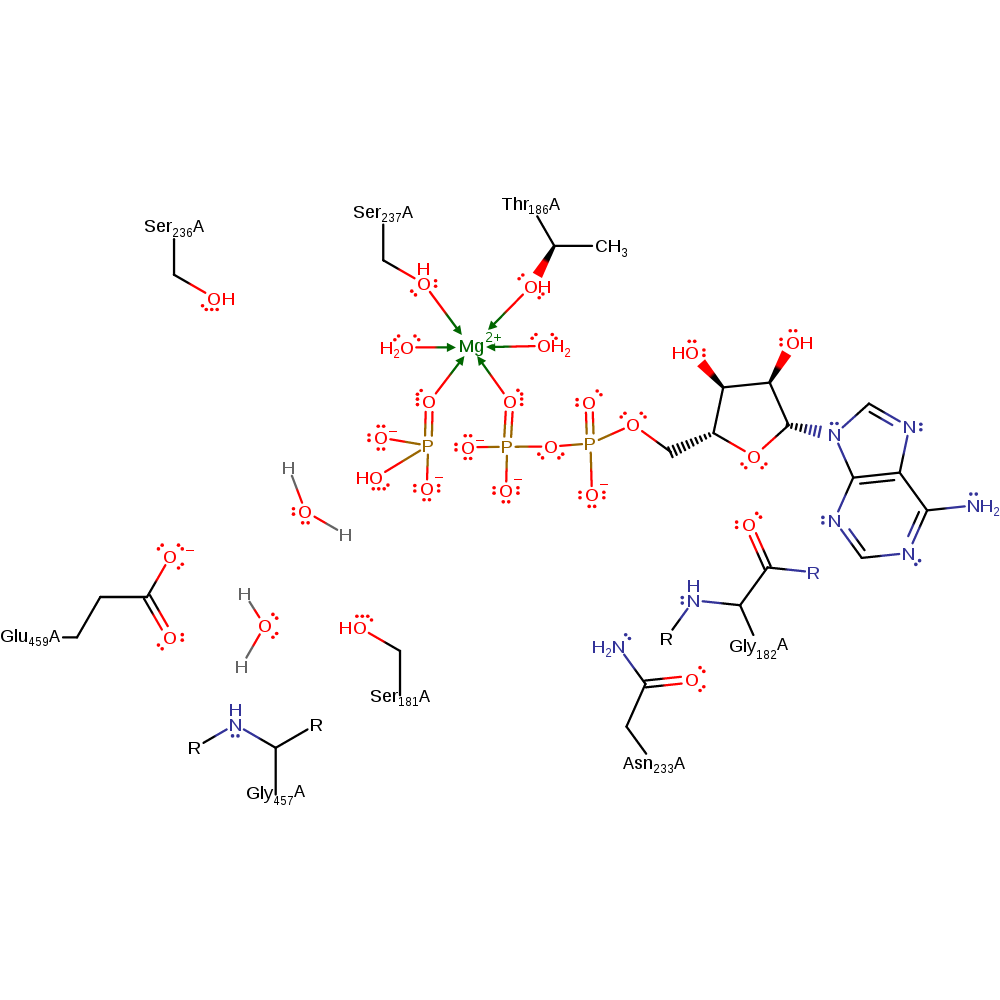 Download:
Download: