Biotin---[acetyl-CoA-carboxylase] ligase
The biotin operon repressor, biotin--[acetyl-CoA-carboxylase] ligase or BirA, is a 33.5-kDa protein. BirA is bifunctional, serving both as the biotin activating enzyme and as a transcriptional regulator. It catalyses the formation of biotinyl-5'-adenylate(bio-5'-AMP) from biotin and ATP and transfers biotin to a specific lysine residue on the biotin carboxyl carrier protein, a subunit of acetyl-CoA carboxylase. Bio-5'-AMP is also the corepressor of BirA.
Reference Protein and Structure
- Sequence
-
P06709
 (6.3.4.15)
(6.3.4.15)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1bib
- THE E. COLI BIOTIN HOLOENZYME SYNTHETASE(SLASH)BIO REPRESSOR CRYSTAL STRUCTURE DELINEATES THE BIOTIN AND DNA-BINDING DOMAINS
(2.8 Å)



- Catalytic CATH Domains
-
3.30.930.10
 (see all for 1bib)
(see all for 1bib)
Enzyme Reaction (EC:6.3.4.15)
Enzyme Mechanism
Introduction
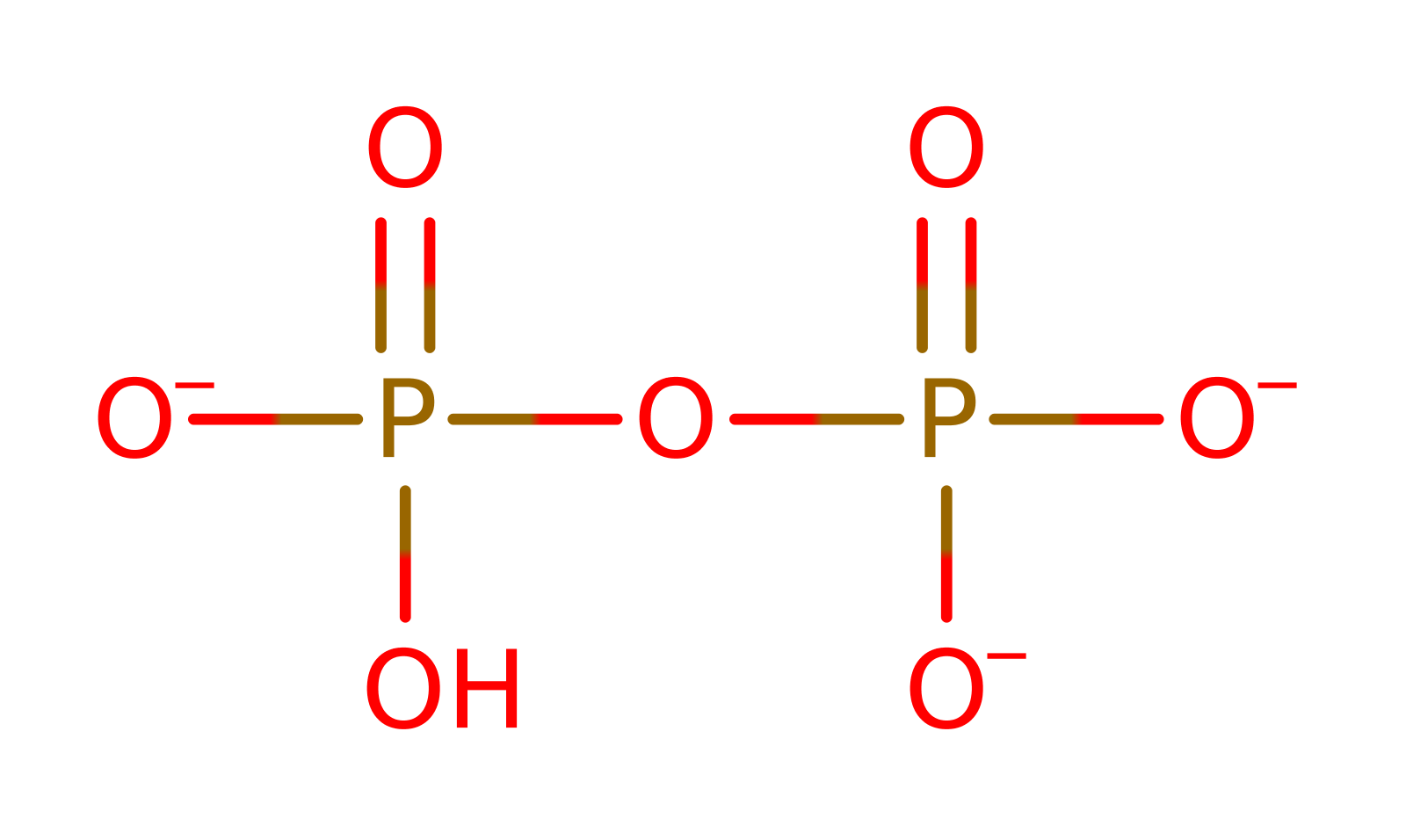
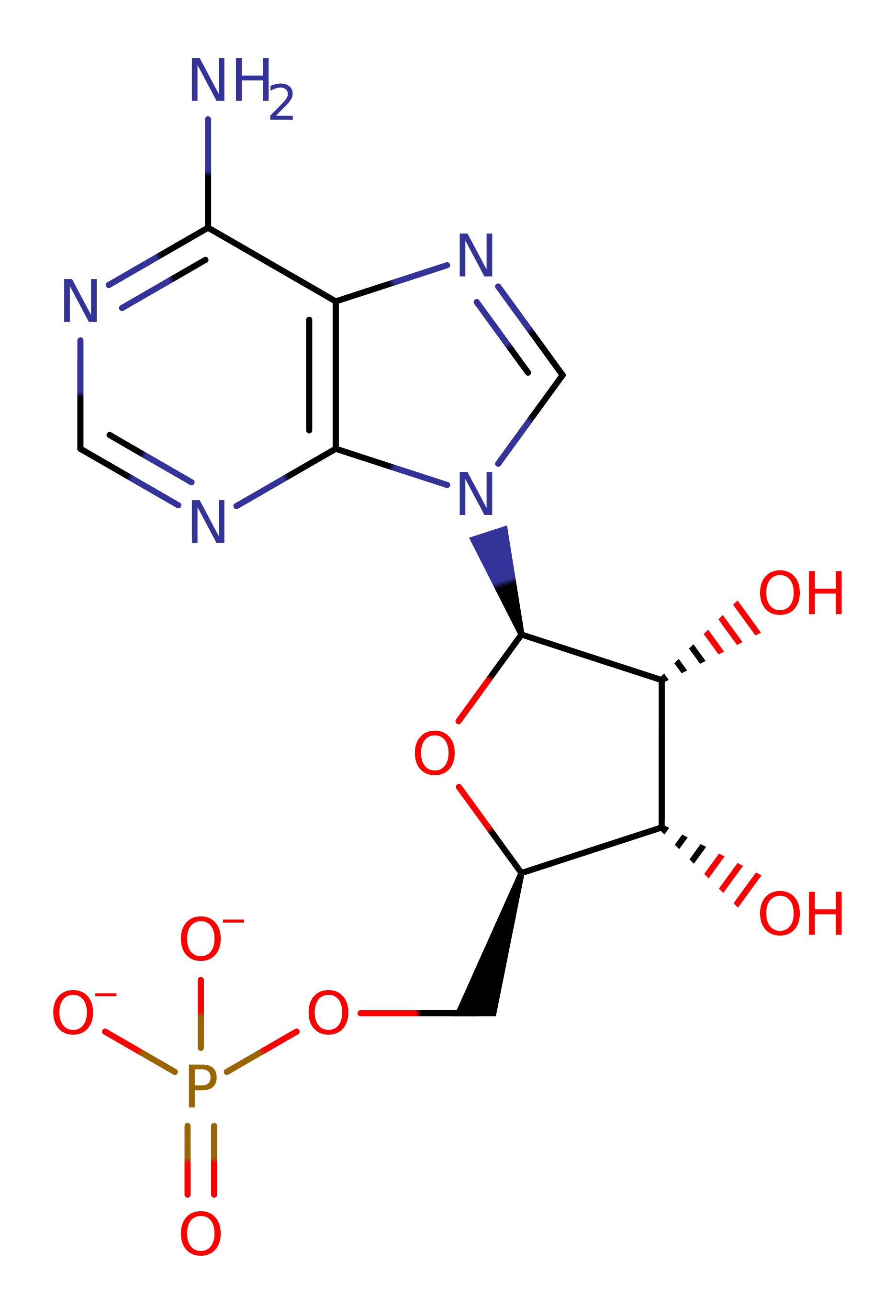
In the first half reaction, Lys183 catalyzes the attack of an oxygen atom of the biotin carboxylate group on P-alpha of ATP to form bio-5'-AMP plus pyrophosphate. The charged transition state is stabilised by a number of arginine residues. Bio-5'-AMP remains bound in the active site and is quite stable. In the presence of the correct apoprotein, apoBCCP, the nucleophilic amino group of the Lys122 to be modified attacks the mixed anhydride carbon atom, thus forming an amide bond between biotin and the lysine side chain with AMP as the other product. Again Lys183 of BirA acts to stabilise the charged transition state. Once the amide bond is formed, the biotin moiety remains attached throughout the lifetime of the protein.
Catalytic Residues Roles
| UniProt | PDB* (1bib) | ||
| Arg118, Arg317 | Arg118A, Arg317A | The positive charge stabilises the negative charges of the phosphate group on ATP. | electrostatic stabiliser |
| Lys183 | Lys183A | The positive charge stabilises the negative charges of BTN carboxylate group and phosphate group. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, intermediate formation, overall reactant used, bimolecular nucleophilic addition, intramolecular elimination, overall product formedReferences
- Bagautdinov B et al. (2008), J Biol Chem, 283, 14739-14750. Protein Biotinylation Visualized by a Complex Structure of Biotin Protein Ligase with a Substrate. DOI:10.1074/jbc.m709116200. PMID:18372281.
- Ma Q et al. (2014), Protein Sci, 23, 932-939. Active site conformational changes upon reaction intermediate biotinyl-5'-AMP binding in biotin protein ligase from Mycobacterium tuberculosis. DOI:10.1002/pro.2475. PMID:24723382.
- Purushothaman S et al. (2008), PLoS One, 3, e2320-. Ligand Specificity of Group I Biotin Protein Ligase of Mycobacterium tuberculosis. DOI:10.1371/journal.pone.0002320. PMID:18509457.
- Wood ZA et al. (2006), J Mol Biol, 357, 509-523. Co-repressor Induced Order and Biotin Repressor Dimerization: A Case for Divergent Followed by Convergent Evolution. DOI:10.1016/j.jmb.2005.12.066. PMID:16438984.
- Bagautdinov B et al. (2005), J Mol Biol, 353, 322-333. Crystal Structures of Biotin Protein Ligase from Pyrococcus horikoshii OT3 and its Complexes: Structural Basis of Biotin Activation. DOI:10.1016/j.jmb.2005.08.032. PMID:16169557.
- Weaver LH et al. (2001), Proc Natl Acad Sci U S A, 98, 6045-6050. Corepressor-induced organization and assembly of the biotin repressor: A model for allosteric activation of a transcriptional regulator. DOI:10.1073/pnas.111128198. PMID:11353844.
- Chapman-Smith A et al. (1999), Trends Biochem Sci, 24, 359-363. The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. PMID:10470036.
- Wilson KP et al. (1992), Proc Natl Acad Sci U S A, 89, 9257-9261. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. DOI:10.1073/pnas.89.19.9257. PMID:1409631.

Step 1. The biotinate and the ATP condense via a nucleophillic substitution this forms the Bionyl-5̕-AMP intermediate and releases pyrophospahte. Lys 183 is essential for this process as it coordinates both the biotinate and the ATP via hydrogen bonding. It also stabilizes the intermediate formed. Arg 118 is also important for coordination, by electrostatic stabilization. There are other arginine residues in the catalytic site which also have some role coordination, such as R317 though 118 is the most relevant.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg118A | electrostatic stabiliser |
| Lys183A | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic substitution, intermediate formation, overall reactant used
Step 2. There is nucleophillic attack from the lysinium residue to the carbon of the mixed anhydride bond of the Bionyl-5̕-AMP intermediate forming a new intermediate with the lysine covalently bonded to this carbon. Lys 183 and Arg 118 and other argines continue to coordinate and stabilize the intermediates via hydrogen bonds and other electrostatic interactions, keeping Bionyl-5̕-AMP in the active site while the while the nucleophillic attack occurs.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg118A | electrostatic stabiliser |
| Lys183A | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation
Step 3. In the final step of the reaction AMP is cleaved from the biotin, leaving the biotin only attached to the lysine by an amide bond where it remains permanently. This overall is a nucleophillic substitution reaction. The intermediate is again stabilized by the hydrogen bonding and electrostatic interactions of the Lys183 and Arg118 and other arginine residues.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg118A | electrostatic stabiliser |
| Lys183A | electrostatic stabiliser |





 Download:
Download: