N-acetylneuraminate lyase
The N-acetylneuraminate lyase (NAL) enzymes are a subfamily of the (beta/alpha)8 enzymes. They all share a common catalytic step but catalyse reactions in different biological pathways. The formation of a Schiff base between a strictly conserved lysine residue and the C2 carbon of the common keto-acid moiety of the substrate is a common feature of the group, with the NAL enzymes catalysing the aldol cleavage of N-acetylneuraminate to form N-acetylmannosine and pyruvate via the distinctive Schiff base intermediate.
Reference Protein and Structure
- Sequence
-
P0A6L4
 (4.1.3.3)
(4.1.3.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1fdy
- N-ACETYLNEURAMINATE LYASE IN COMPLEX WITH HYDROXYPYRUVATE
(2.45 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1fdy)
(see all for 1fdy)
Enzyme Reaction (EC:4.1.3.3)
Enzyme Mechanism
Introduction
The initial step is thought to be ring opening of the substrate alpha anomer. The conserved Lys165 then acts as nucleophile to the carbonyl group with concomitant proton transfer from the Lys165 nitrogen to the alcohol group of the tetrahedral intermediate. A second proton transfer results in a cationic oxonium group. The attacking nitrogen lone pair then kicks out a water molecule forming the substrate-enzyme Schiff base intermediate. A tyrosine group mediates the proton abstraction from the hydroxyl at the 4 position by the carboxylate group, leading to the formation of an enamine. Protonation at the methylene carbon then converts the enamine into an imine. Subsequent ring closure of N-acetylmannosine is spontaneous. The collapse of the enzyme-substrate intermediate is brought about by a hydrolytic water molecule which leads to the liberation of Lys165 and pyruvate.
Catalytic Residues Roles
| UniProt | PDB* (1fdy) | ||
| Lys165 | Lys165A | Forms a Schiff-base intermediate with the substrate. Also involved in ring opening. | covalently attached, nucleofuge, nucleophile, proton acceptor, electron pair acceptor, electron pair donor |
| Tyr137 | Tyr137A | Involved in proton transfer during cleavage. | proton relay, proton acceptor, proton donor |
Chemical Components
decyclisation, proton transfer, overall reactant used, intermediate formation, bimolecular nucleophilic addition, intramolecular elimination, schiff base formed, bimolecular elimination, overall product formed, proton relay, cyclisation, tautomerisation (not keto-enol), intermediate terminated, native state of enzyme regeneratedReferences
- Lawrence MC et al. (1997), J Mol Biol, 266, 381-399. Structure and mechanism of a sub-family of enzymes related to N-acetylneuraminate lyase. DOI:10.1006/jmbi.1996.0769. PMID:9047371.
- Daniels AD et al. (2014), ACS Chem Biol, 9, 1025-1032. Reaction mechanism of N-acetylneuraminic acid lyase revealed by a combination of crystallography, QM/MM simulation, and mutagenesis. DOI:10.1021/cb500067z. PMID:24521460.
- Barbosa JA et al. (2000), J Mol Biol, 303, 405-421. Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme. DOI:10.1006/jmbi.2000.4138. PMID:11031117.

Step 1. The reaction begins with the ring opening. Which is promoted by Lys165.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys165A | proton acceptor |
Chemical Components
decyclisation, proton transfer
Step 2. Lys165 performs a nucleophilic attack upon C2 forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys165A | covalently attached, nucleophile |
Chemical Components
overall reactant used, intermediate formation, ingold: bimolecular nucleophilic addition, proton transfer
Step 3. Water is eliminated from this intermediate and an imine is formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys165A | covalently attached, electron pair donor |
Chemical Components
ingold: intramolecular elimination, schiff base formed
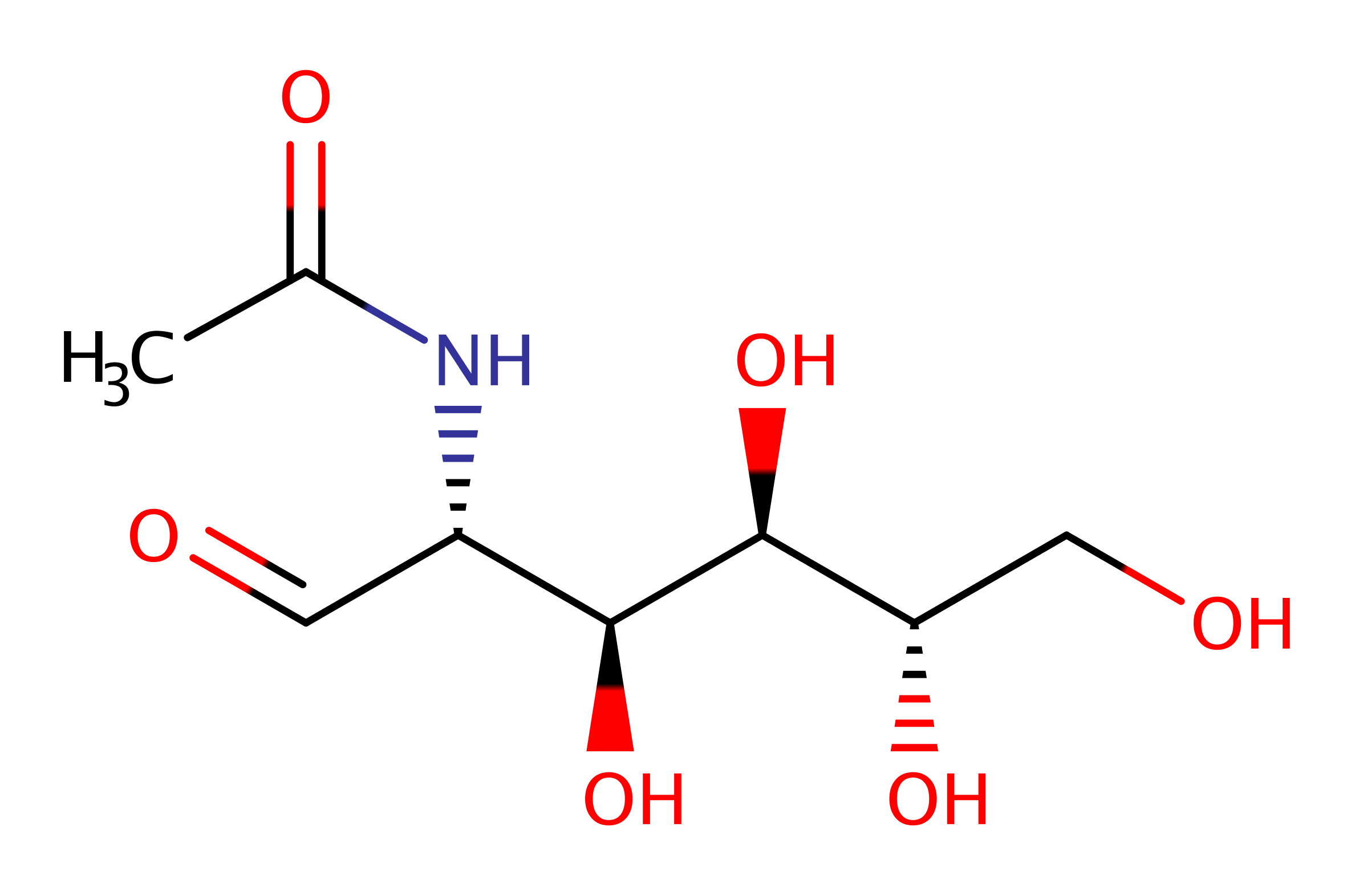
Step 4. The bond between C3 and C4 is broken releasing the linear form of the product aldehydo-N-acetyl-D-mannosamine and an enamine intermediate is formed. This is assisted by Tyr143 which acts as a proton relay.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys165A | covalently attached |
| Tyr137A | proton relay, proton donor |
| Lys165A | electron pair acceptor |
| Tyr137A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, overall product formed, proton relay
Step 5. Aldehydo-N-acetyl-D-mannosamine is converted to its cyclic form. The enamine tautomerizes forming a new imine intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys165A | covalently attached, electron pair donor |
Chemical Components
proton transfer, cyclisation, tautomerisation (not keto-enol), schiff base formed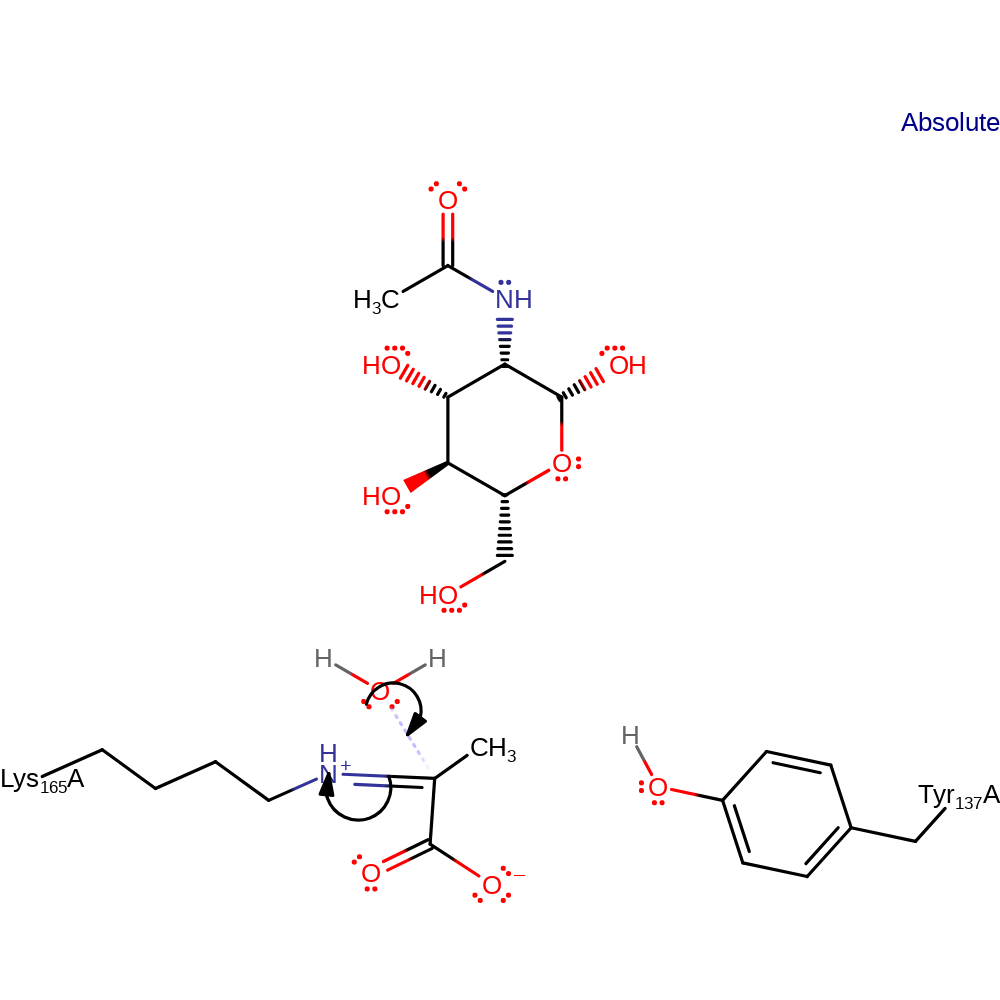
Step 6. A hydrolytic water performs a nucleophilic attack upon the imine carbon forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys165A | covalently attached, electron pair acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition
Step 7. Lys165 is eliminated from the intermediate and the other product pyruvate is formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys165A | nucleofuge |


 Download:
Download: