ADP-glyceromanno-heptose 6-epimerase
ADP -L-glycero-D-mannoheptose 6-epimerase (AGME) is classified as a member of the short-chain dehydrogenase/reductase (SDR) superfamily, on the grounds of its high structural similarity with UDP-galactose epimerase and its positioning of conserved catalytic residues. AGME catalyses the interconversion between ADP-D-glycero-D-mannoheptose and ADP-L-glycero-D-mannoheptose, the last step in the biosynthesis of the precursor of L-glycero-D-mannoheptose. The enzyme is an NADP dependent epimerase which shows a diminished activity with NAD.
Reference Protein and Structure
- Sequence
-
P67910
 (5.1.3.20)
(5.1.3.20)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1eq2
- THE CRYSTAL STRUCTURE OF ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE
(2.0 Å)



- Catalytic CATH Domains
-
3.90.25.10
 3.40.50.720
3.40.50.720  (see all for 1eq2)
(see all for 1eq2)
- Cofactors
- Nadp zwitterion (1)
Enzyme Reaction (EC:5.1.3.20)
Enzyme Mechanism
Introduction
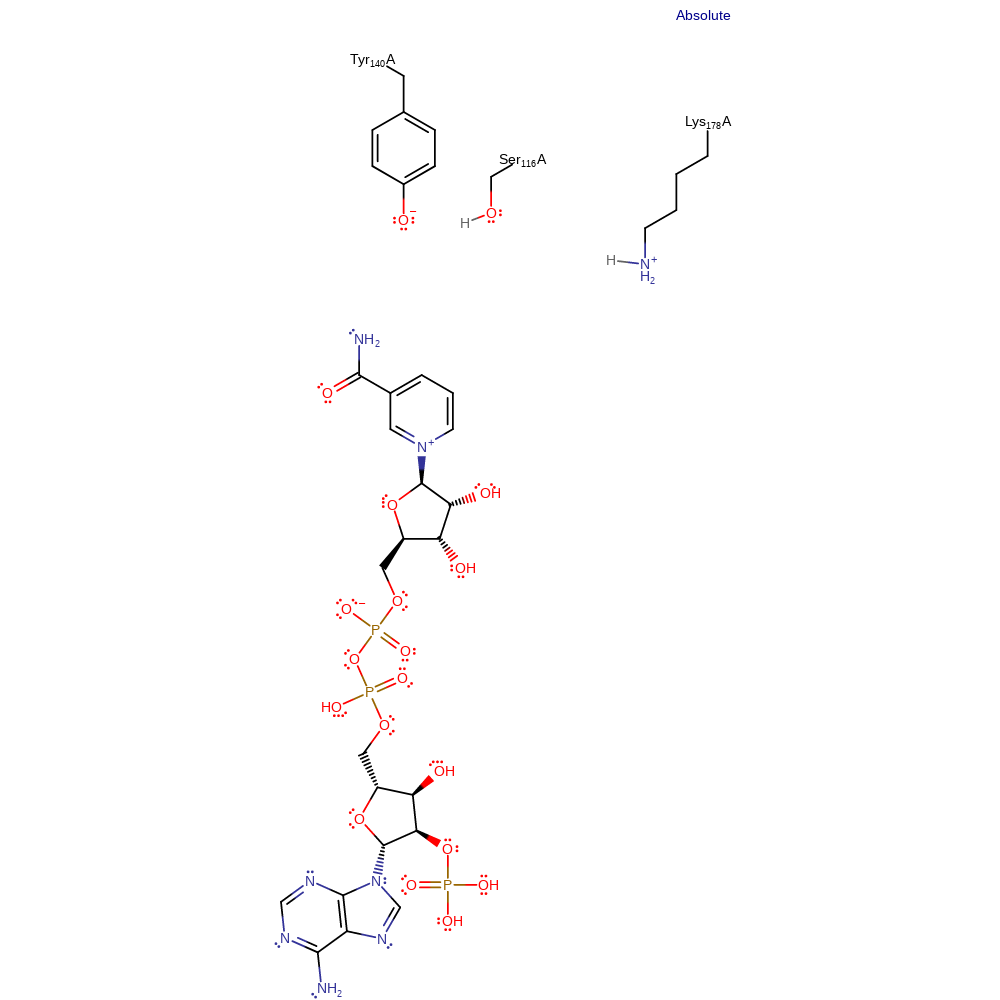
This is the two-base mechanism in which both the tyrosine and lysine act as general acid/bases. The basic mechanism is identical to the two-base proposal: the enzyme bound NAD+ coenzyme accepts a hydride from the substrate's C-6. Hydride transfer is promoted by deprotonation at C6-OH. The resulting keto intermediate flips in the active site to allow return of the hydride from NADH to the other face of the substrate, forming the alternative epimer. The enzyme is able to catalyse both the forward and reverse reactions, with separate catalytic residues acting as the base in each direction.
Catalytic Residues Roles
| UniProt | PDB* (1eq2) | ||
| Lys178 | Lys178A | Acts as a general base towards the ADP-L-glycero-D-mannoheptose substrate in the reverse reaction, removing the C6-OH proton, facilitating hydride transfer to NAD+ cofactor. | proton acceptor, proton donor |
| Tyr140 | Tyr140A | The residue acts as a general base towards the C6-OH hydrogen of ADP-D-glycero-D-mannoheptose, facilitating the removal of hydride to the NAD cofactor via a proton shuttle with Ser 116. | proton acceptor, proton donor |
| Ser116 | Ser116A | The residue acts as a proton shuttle between the sugar substrate and the phenolic side chain of Tyr 140. | proton relay, proton acceptor, proton donor |
Chemical Components
proton transfer, hydride transfer, overall reactant used, native state of cofactor regenerated, overall product formed, inferred reaction step, native state of enzyme regeneratedReferences
- Morrison JP et al. (2007), Biochemistry, 46, 3916-3924. A Two-Base Mechanism forEscherichia coliADP-l-glycero-d-manno-Heptose 6-Epimerase†. DOI:10.1021/bi602641m. PMID:17316025.
- Kowatz T et al. (2010), Protein Sci, 19, 1337-1343. The crystal structure of the Y140F mutant of ADP-L-glycero-D-manno-heptose 6-epimerase bound to ADP-beta-D-mannose suggests a one base mechanism. DOI:10.1002/pro.410. PMID:20506248.
- Mayer A et al. (2007), Biochemistry, 46, 6149-6155. Intermediate release by ADP-L-glycero-D-manno-heptose 6-epimerase. DOI:10.1021/bi700332h. PMID:17455913.
- Morrison JP et al. (2005), Biochemistry, 44, 5907-5915. Dismutase activity of ADP-L-glycero-D-manno-heptose 6-epimerase: evidence for a direct oxidation/reduction mechanism. DOI:10.1021/bi050106c. PMID:15823050.
- Ni Y et al. (2001), J Biol Chem, 276, 27329-27334. Evidence that NADP+ is the physiological cofactor of ADP-L-glycero-D-mannoheptose 6-epimerase. DOI:10.1074/jbc.M102258200. PMID:11313358.
- Deacon AM et al. (2000), Structure, 8, 453-462. The crystal structure of ADP-l-glycero-d-mannoheptose 6-epimerase: catalysis with a twist. DOI:10.1016/s0969-2126(00)00128-3. PMID:10896473.
- Thoden JB et al. (1996), Protein Sci, 5, 2149-2161. High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol. DOI:10.1002/pro.5560051102. PMID:8931134.

Step 1. Tyr140 abstracts a proton from the sugar substrate via Ser116. This in turn eliminates the hydride to NADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr140A | proton acceptor |
| Ser116A | proton donor, proton relay, proton acceptor |
Chemical Components
proton transfer, hydride transfer, overall reactant used
Step 2. The newly formed carbonyl group rotates in the active site and the NADP returns the hydride to the intermediate with concomitant deprotonation of Lys178.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys178A | proton donor |
Chemical Components
native state of cofactor regenerated, proton transfer, hydride transfer, overall product formed
Step 3. Inferred step in which the Lys178 abstracts a proton from Tyr140 via Ser116 to regenerate the active site ground state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser116A | proton acceptor |
| Tyr140A | proton donor |
| Lys178A | proton acceptor |
| Ser116A | proton donor, proton relay |
Chemical Components
inferred reaction step, proton transfer, native state of enzyme regeneratedIntroduction
This is the one-base mechanism in which the tyrosine acts as the only general acid/base. The basic mechanism is identical to the two-base proposal: the enzyme bound NAD+ coenzyme accepts a hydride from the substrate's C-6. Hydride transfer is promoted by deprotonation at C6-OH. The resulting keto intermediate flips in the active site to allow return of the hydride from NADH to the other face of the substrate, forming the alternative epimer. The enzyme is able to catalyse both the forward and reverse reactions.
Catalytic Residues Roles
| UniProt | PDB* (1eq2) | ||
| Lys178 | Lys178A | Acts to stabilise the reactive intermediates and transition states formed during the course of the reaction. | electrostatic stabiliser |
| Tyr140 | Tyr140A | The residue acts as a general base towards the C6-OH hydrogen of ADP-D-glycero-D-mannoheptose, facilitating the removal of hydride to the NAD cofactor. It also acts as the general acid in the final step of the reaction. | proton acceptor, proton donor |
| Ser116 | Ser116A | Helps stabilise and activate the negatively charged catalytic tyrosine residue. | electrostatic stabiliser |
Chemical Components
overall reactant used, hydride transfer, proton transfer, overall product formed, native state of cofactor regenerated, native state of enzyme regeneratedReferences
- Kowatz T et al. (2010), Protein Sci, 19, 1337-1343. The crystal structure of the Y140F mutant of ADP-L-glycero-D-manno-heptose 6-epimerase bound to ADP-beta-D-mannose suggests a one base mechanism. DOI:10.1002/pro.410. PMID:20506248.

Step 1. Tyr140 abstracts a proton from the sugar substrate via Ser116. This in turn eliminates the hydride to NADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser116A | electrostatic stabiliser |
| Lys178A | electrostatic stabiliser |
| Tyr140A | proton acceptor |
Chemical Components
overall reactant used, hydride transfer, proton transfer
Step 2. The newly formed carbonyl group rotates in the active site and the NADP returns the hydride to the intermediate with concomitant deprotonation of Tyr140.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser116A | electrostatic stabiliser |
| Lys178A | electrostatic stabiliser |
| Tyr140A | proton donor |


 Download:
Download:  Download:
Download: