Cellulase (GH9)
Cellulase E4 from Thermomonospora is both exo- and endo- cellulases, catalysing the hydrolysis of cellulose. It contains both a family 9 catalytic domain, exhibiting an (alpha/apha)6 barrel fold and a family III cellulose binding domain, having an antiparallel beta-sandwich fold.
Reference Protein and Structure
- Sequence
-
P26221
 (3.2.1.4)
(3.2.1.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermobifida fusca (Bacteria)

- PDB
-
1js4
- ENDO/EXOCELLULASE:CELLOBIOSE FROM THERMOMONOSPORA
(2.0 Å)



- Catalytic CATH Domains
-
1.50.10.10
 (see all for 1js4)
(see all for 1js4)
Enzyme Reaction (EC:3.2.1.4)
Enzyme Mechanism
Introduction
Same as the other members of family 9 cellulase, E4 endo/exocellulase cleaves cullulose with inversion of configuration at the anomeric carbon. In the mechanism, an acid protonates the glycosidic oxygen and a base extracts a nucleophilic water that attacks the anomeric carbon. Here, Glu470 is the acid. Both Asp 104 and Asp 101 hydrogen bond to the attacking water and both of them appear to contribute to the activation of the water molecule. Asp 104 appears better oriented to accept a proton from the water than Asp 101, however both residues may be able to act as the base.
Catalytic Residues Roles
| UniProt | PDB* (1js4) | ||
| Glu470 | Glu424A | It acts as an acid to protonate the glycosidic oxygen. | promote heterolysis, proton acceptor, proton donor |
| Tyr252 | Tyr206A | Hydrogen bonds to be Asp101 contributing to the activation of water. | electrostatic stabiliser |
| Asp104 | Asp58A | It deprotonates the water molecule, which nucleophillically attacks the glycosidic bond of the substrate in hydrolysis. | activator, increase nucleophilicity, proton acceptor, proton donor |
| Asp101 | Asp55A | It forms a hydrogen bond to the water molecule, which nucleophillically attacks the glycosidic bond of the substrate in hydrolysis, to activate it. Some evidence suggests this residue may be also be the base. | activator, increase nucleophilicity, electrostatic stabiliser |
Chemical Components
hydrolysis, overall product formed, overall reactant used, bimolecular nucleophilic substitution, proton transfer, inferred reaction step, native state of enzyme regeneratedReferences
- Sakon J et al. (1997), Nat Struct Biol, 4, 810-818. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. DOI:10.1038/nsb1097-810. PMID:9334746.
- Zhou W et al. (2004), Biochemistry, 43, 9655-9663. Kinetic studies of Thermobifida fusca Cel9A active site mutant enzymes. DOI:10.1021/bi049394n. PMID:15274620.
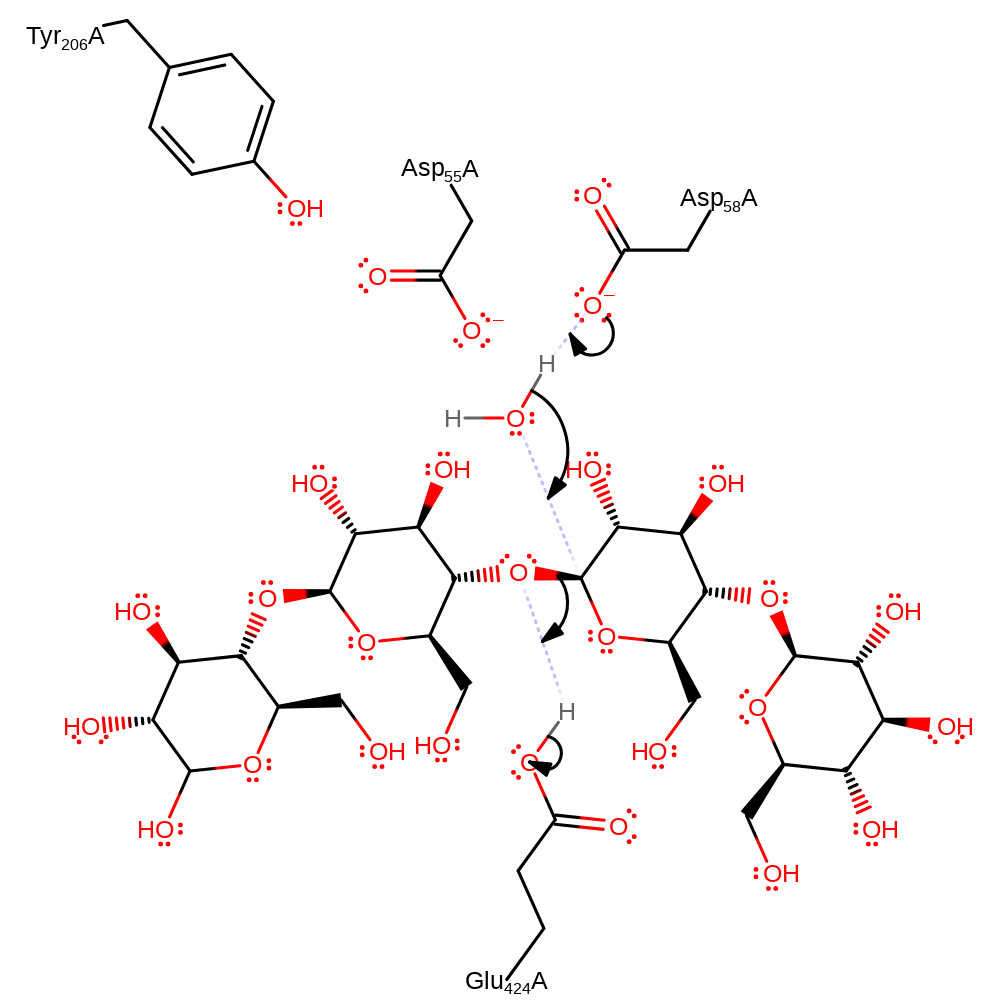
Step 1. Either Asp101 or Asp104 deprotonates the water activating it for nucleophilic attack on the anomeric carbon. (Tyr252 hydrogen bonds to Asp101 which contributes to the activation of water.) This causes the glycosidic bond to be broken upon protonation by Glu470.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp55A | electrostatic stabiliser, activator |
| Asp58A | activator |
| Tyr206A | electrostatic stabiliser |
| Asp55A | increase nucleophilicity |
| Asp58A | increase nucleophilicity |
| Glu424A | promote heterolysis |
| Asp58A | proton acceptor |
| Glu424A | proton donor |
Chemical Components
hydrolysis, overall product formed, overall reactant used, ingold: bimolecular nucleophilic substitution, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu424A | proton acceptor |
| Asp58A | proton donor |




 Download:
Download: