3-dehydroquinate synthase
Dehydroquinate synthase (DHQS) has long been regarded as a catalytic wonder due to its ability to perform several consecutive chemical reactions in one active site. DHQS performs the second step in the shikimate pathway, which is required for the synthesis of aromatic compounds in bacteria, microbial eukaryotes and plants. The fold of the C-terminal domain of DHQS shows no detectable structural relationships to known structures. The active site is located between the two domains. The C-terminal domain contains most of the residues involved in catalysis and in substrate and Zn(2+) binding. The crystal structure of DHQS shows an active site which possesses a variety of interactions between metal, dinucleotide, substrate analogue and protein.
Reference Protein and Structure
- Sequence
-
P07547
 (1.1.1.25, 2.5.1.19, 2.7.1.71, 4.2.1.10, 4.2.3.4)
(1.1.1.25, 2.5.1.19, 2.7.1.71, 4.2.1.10, 4.2.3.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Aspergillus nidulans FGSC A4 (Fungus)

- PDB
-
1dqs
- CRYSTAL STRUCTURE OF DEHYDROQUINATE SYNTHASE (DHQS) COMPLEXED WITH CARBAPHOSPHONATE, NAD+ AND ZN2+
(1.8 Å)



- Catalytic CATH Domains
-
1.20.1090.10
 3.40.50.1970
3.40.50.1970  (see all for 1dqs)
(see all for 1dqs)
- Cofactors
- Zinc(2+) (1), Water (2), Nadph(4-) (1), Nadp zwitterion (1) Metal MACiE
Enzyme Reaction (EC:4.2.3.4)
Enzyme Mechanism
Introduction
General base catalysis is prompted by His275, which activates the water molecule (Wat7) to lose its hydrogen. This activated water then deprotonates the C5 hydroxyl of DHAP. In concert, the C5 oxygen forms a carbonyl and the C5 hydride is transferred to the C4 position of NAD+. A Zn(II) ion facilitates this step by polarising the hydroxyl group and stabilising the transition state. The phosphate of the intermediate removes the C6 proton, leading to the beta-elimination of the phosphate group. NADH transfers a hydride to the C5 position and the developing alkoxide is protonated by a water molecule, which is in turn protonated by His275. Zn(II) facilitates this reaction by polarising the carbonyl and stabilising the transient negative charge on the oxygen. A second general base catalysis is activated by an unknown base, possibly Arg260, accepting a proton from Wat20 and this is then stabilised by Asn268. Lys152 then stabilises the open ring, which reforms after a rotation about the C5-C6 bond and an intramolecular aldol condensation.
Catalytic Residues Roles
| UniProt | PDB* (1dqs) | ||
| His275 | His275A | Part of a proton-shuttling system. It deprotonates water during DHAP oxidation and protonates water during the reduction of the intermediate. It is also potentially involved in the beta-elimination of the phosphate by stabilising the transition state and/or forcing the phosphate to adopt the correct position for C6 proton abstraction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Lys152 | Lys152A | Forms part of the K152/N162 "pincer", one of the key interactions in ensuring that the active site is rigid. Forms part of the phosphate binding pocket of the substrate. It is also implicated in preventing epimeriaation at C2 by interacting with the carboxylic acid group on that carbon. Finally, it has been implicated in the stabilisation of the negative charge generated during the final intramolecular aldol condensation. | hydrogen bond donor, steric role |
| Glu194, His271, His287 | Glu194A, His271A, His287A | Forms part of the catalytic zinc binding site. | metal ligand |
| Arg264, Asn268 | Arg264A, Asn268A | Forms part of the R264/N268 "pincer", one of the key interactions in ensuring that the active site is rigid. | hydrogen bond donor, electrostatic stabiliser, steric role |
| Arg130 | Arg130B | Stabilises the base of the closed form of the active site by cross-linking to the rigid core domain. Also involved in the binding and stabilisation of the substrate. | electrostatic stabiliser |
| Asn268, Lys250 | Asn268A, Lys250A | Involved in ensuring that the reaction intermediates in steps four and five are held in the correct position to prevent the epimerisation of the substrate. | hydrogen bond donor, steric role |
| Glu260 | Glu260A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
Chemical Components
bimolecular elimination, aromatic bimolecular nucleophilic addition, hydride transfer, intermediate formation, overall reactant used, cofactor used, proton transfer, intramolecular elimination, overall product formed, dephosphorylation, intermediate collapse, aromatic unimolecular elimination by the conjugate base, bimolecular nucleophilic addition, proton relay, native state of cofactor regenerated, decyclisation, charge delocalisation, intramolecular nucleophilic addition, aldol addition, cyclisation, intermediate terminated, native state of enzyme regeneratedReferences
- Carpenter EP et al. (1998), Nature, 394, 299-302. Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis. DOI:10.1038/28431. PMID:9685163.
- Mittelstädt G et al. (2013), Arch Biochem Biophys, 537, 185-191. Biochemical and structural characterisation of dehydroquinate synthase from the New Zealand kiwifruit Actinidia chinensis. DOI:10.1016/j.abb.2013.07.022. PMID:23916589.
- de Mendonça JD et al. (2011), Mol Biosyst, 7, 119-128. Kinetic mechanism determination and analysis of metal requirement of dehydroquinate synthase from Mycobacterium tuberculosisH37Rv: an essential step in the function-based rational design of anti-TB drugs. DOI:10.1039/c0mb00085j. PMID:20978656.
- Negron L et al. (2011), Org Biomol Chem, 9, 2861-. Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by dehydroquinate synthase. DOI:10.1039/c0ob01141j. PMID:21387027.
- Liu JS et al. (2008), Biochem Biophys Res Commun, 373, 1-7. Structure-based inhibitor discovery of Helicobacter pylori dehydroquinate synthase. DOI:10.1016/j.bbrc.2008.05.070. PMID:18503755.
- Sugahara M et al. (2005), Proteins, 58, 249-252. Crystal structure of dehydroquinate synthase from Thermus thermophilus HB8 showing functional importance of the dimeric state. DOI:10.1002/prot.20281. PMID:15508124.
- Park A et al. (2004), Protein Sci, 13, 2108-2119. Biophysical and kinetic analysis of wild-type and site-directed mutants of the isolated and native dehydroquinate synthase domain of the AROM protein. DOI:10.1110/ps.04705404. PMID:15273308.
- Nichols CE et al. (2003), J Mol Biol, 327, 129-144. Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase. DOI:10.1016/s0022-2836(03)00086-x. PMID:12614613.
- Chandran SS et al. (2001), Bioorg Med Chem Lett, 11, 1493-1496. Aromatic inhibitors of dehydroquinate synthase: synthesis, evaluation and implications for gallic acid biosynthesis. DOI:10.1016/s0960-894x(01)00065-8.
- Montchamp J et al. (1997), J Am Chem Soc, 119, 7645-7653. Cyclohexenyl and Cyclohexylidene Inhibitors of 3-Dehydroquinate Synthase: Active Site Interactions Relevant to Enzyme Mechanism and Inhibitor Design. DOI:10.1021/ja961771z.
- Safar, M. et al. (1978), Nouv Presse Med, 7, 2767-2768. [Value of a single oral dose of pindolol]. PMID:IPR030960.

Step 1. His275 (activated by water) deprotonates a water molecule, which in turn deprotonates the alcohol of the sugar substrate at the C5 position. This forms a ketone group, and causes elimination of a hydride ion, which adds to NAD+.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys250A | hydrogen bond donor, electrostatic stabiliser |
| Arg264A | hydrogen bond donor, electrostatic stabiliser |
| Asn268A | hydrogen bond donor |
| His275A | hydrogen bond acceptor, hydrogen bond donor |
| Lys152A | hydrogen bond donor |
| Glu260A | hydrogen bond acceptor |
| Glu194A | metal ligand |
| His287A | metal ligand |
| His271A | metal ligand |
| Arg130B | electrostatic stabiliser |
| His275A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, ingold: aromatic bimolecular nucleophilic addition, hydride transfer, intermediate formation, overall reactant used, cofactor used, proton transfer
Step 2. The phosphate deprotonates the C4 position, causing self elimination.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His275A | hydrogen bond donor, hydrogen bond acceptor |
| Lys250A | hydrogen bond donor, electrostatic stabiliser |
| Arg264A | hydrogen bond donor, electrostatic stabiliser |
| Asn268A | hydrogen bond donor |
| Glu260A | hydrogen bond acceptor |
| Lys152A | hydrogen bond donor |
| Glu194A | metal ligand |
| His287A | metal ligand |
| His271A | metal ligand |
| Arg130B | electrostatic stabiliser |
Chemical Components
ingold: intramolecular elimination, overall product formed, dephosphorylation, intermediate collapse, intermediate formation
Step 3. NADH eliminates a hydride ion, which adds to the C5 position of the sugar ring, causing the ketone to be reduced and deprotonates water, which deprotonates the His275.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His275A | hydrogen bond acceptor, hydrogen bond donor |
| Lys250A | hydrogen bond donor, electrostatic stabiliser |
| Arg264A | hydrogen bond donor, electrostatic stabiliser |
| Asn268A | hydrogen bond donor |
| Glu260A | hydrogen bond acceptor |
| Glu194A | metal ligand |
| His287A | metal ligand |
| His271A | metal ligand |
| His275A | proton donor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: bimolecular nucleophilic addition, hydride transfer, proton relay, native state of cofactor regenerated, intermediate formation, proton transfer
Step 4. Glu260 deprotonates water, which deprotonates the C2 alcohol of the sugar, cleaving the ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys250A | hydrogen bond donor, electrostatic stabiliser, steric role |
| Arg264A | hydrogen bond donor, electrostatic stabiliser, steric role |
| Asn268A | hydrogen bond donor, steric role |
| Glu260A | hydrogen bond acceptor |
| Lys152A | steric role |
| His275A | hydrogen bond acceptor, hydrogen bond donor |
| Glu194A | metal ligand |
| His287A | metal ligand |
| His271A | metal ligand |
| Glu260A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, decyclisation, proton relay, charge delocalisation, intermediate formation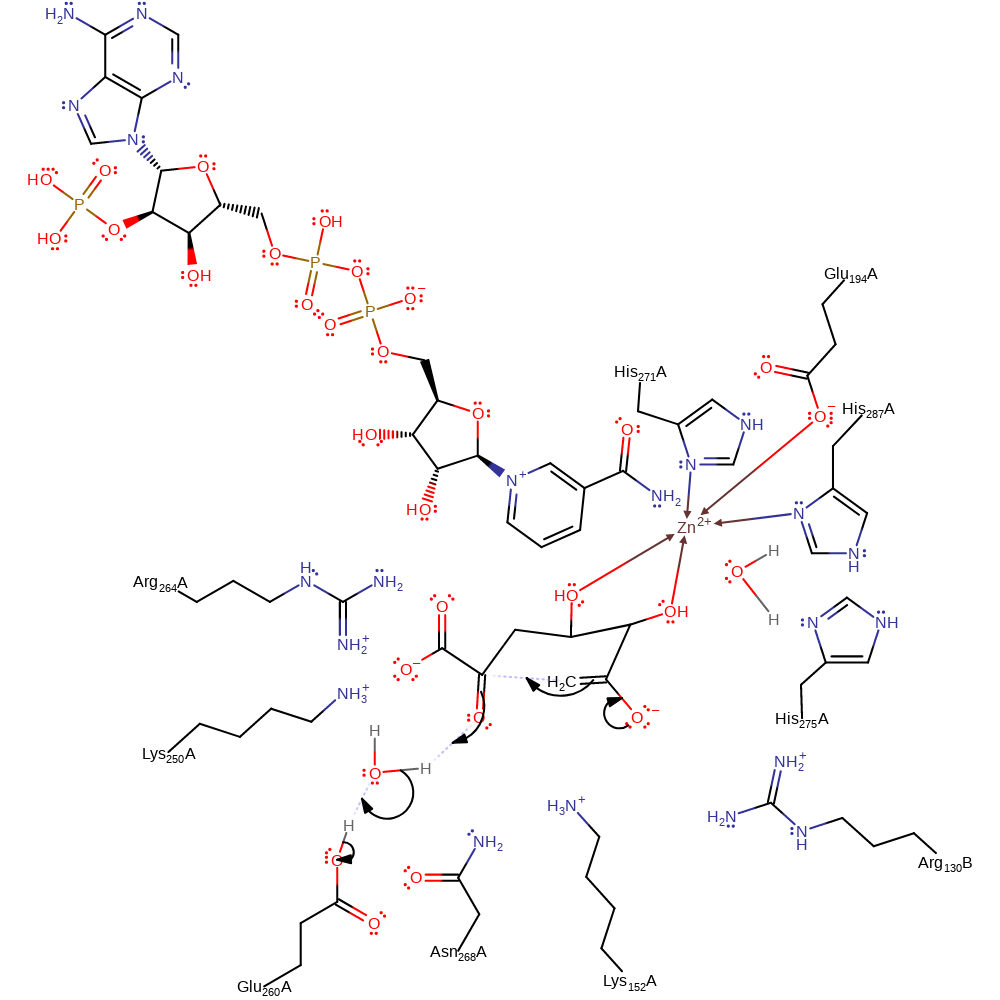
Step 5. The newly formed oxyanion initiates double bond rearrangement, and causes an intramolecular nucleophilic attack on the keto-group formed previously, reforming the ring structure.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu260A | hydrogen bond donor |
| Lys250A | hydrogen bond donor, electrostatic stabiliser, steric role |
| Arg264A | hydrogen bond donor, electrostatic stabiliser, steric role |
| Asn268A | hydrogen bond donor, steric role |
| Lys152A | steric role |
| His275A | hydrogen bond acceptor, hydrogen bond donor |
| Glu194A | metal ligand |
| His287A | metal ligand |
| His271A | metal ligand |
| Glu260A | proton donor |



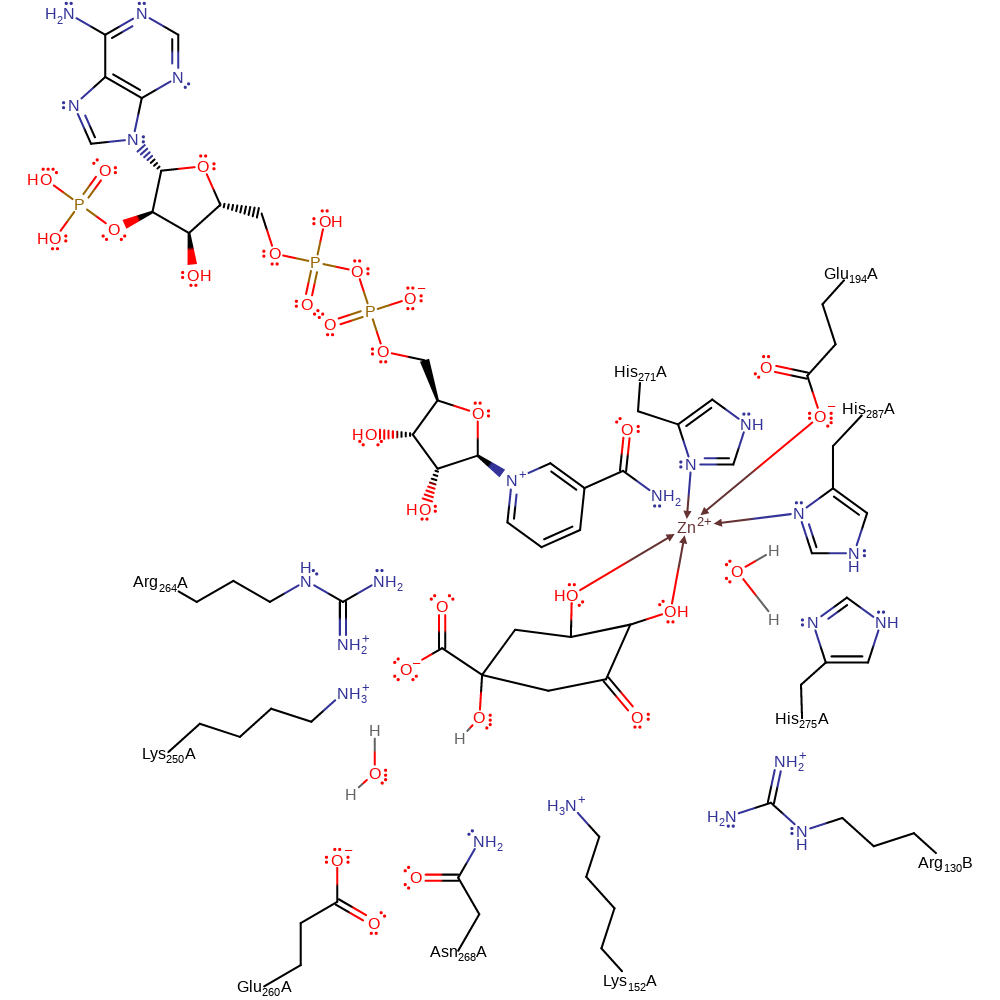 Download:
Download: