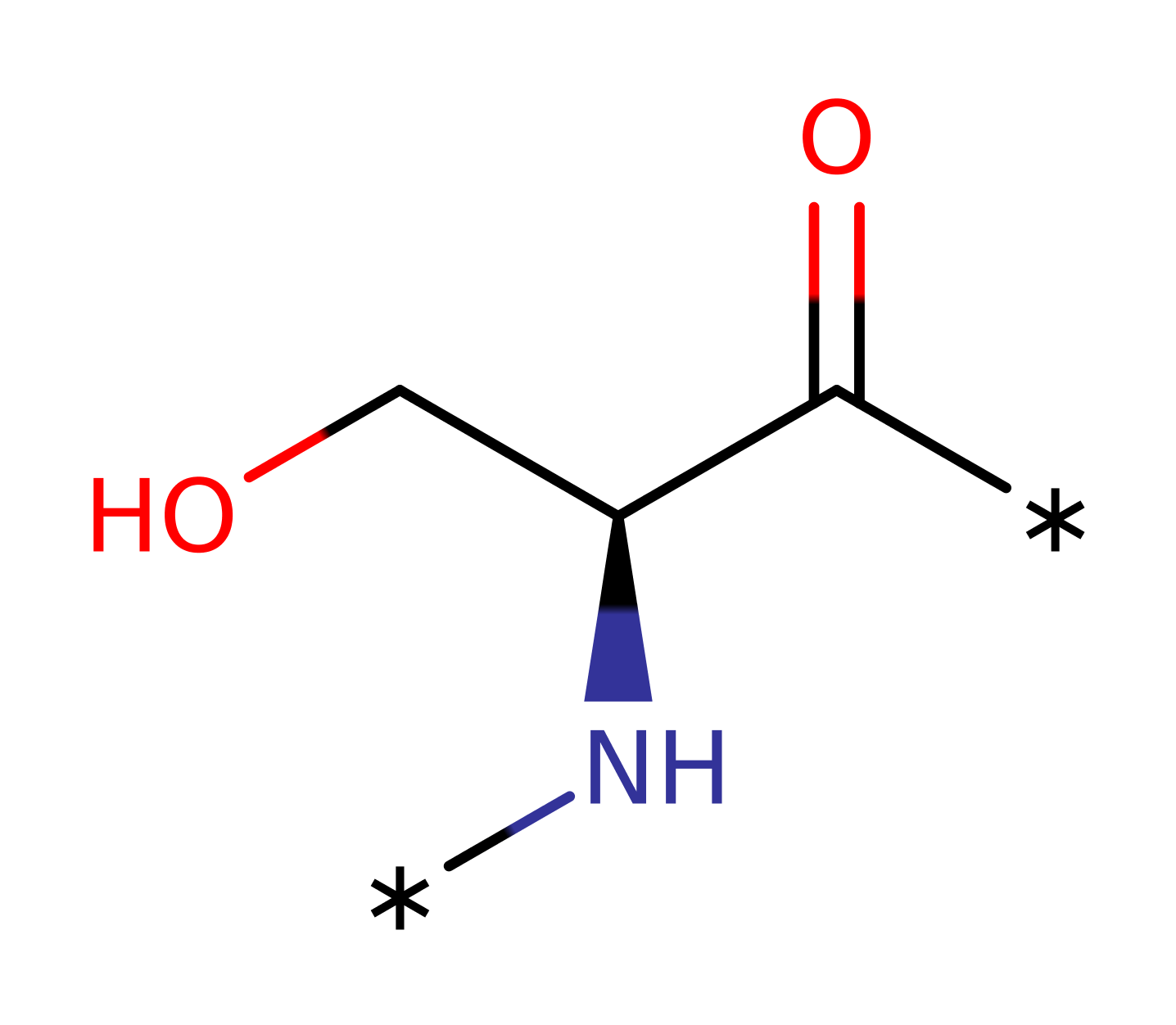
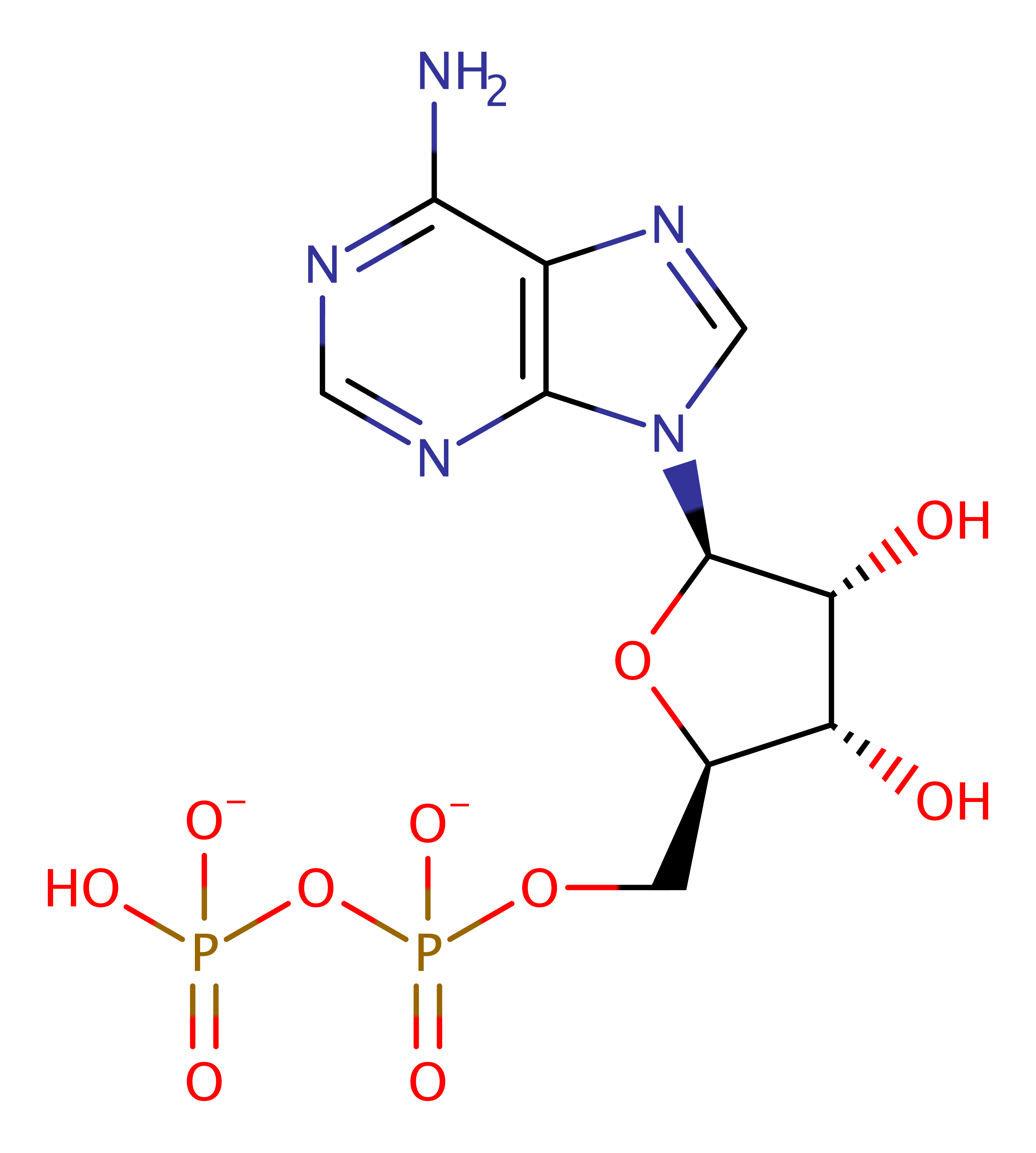
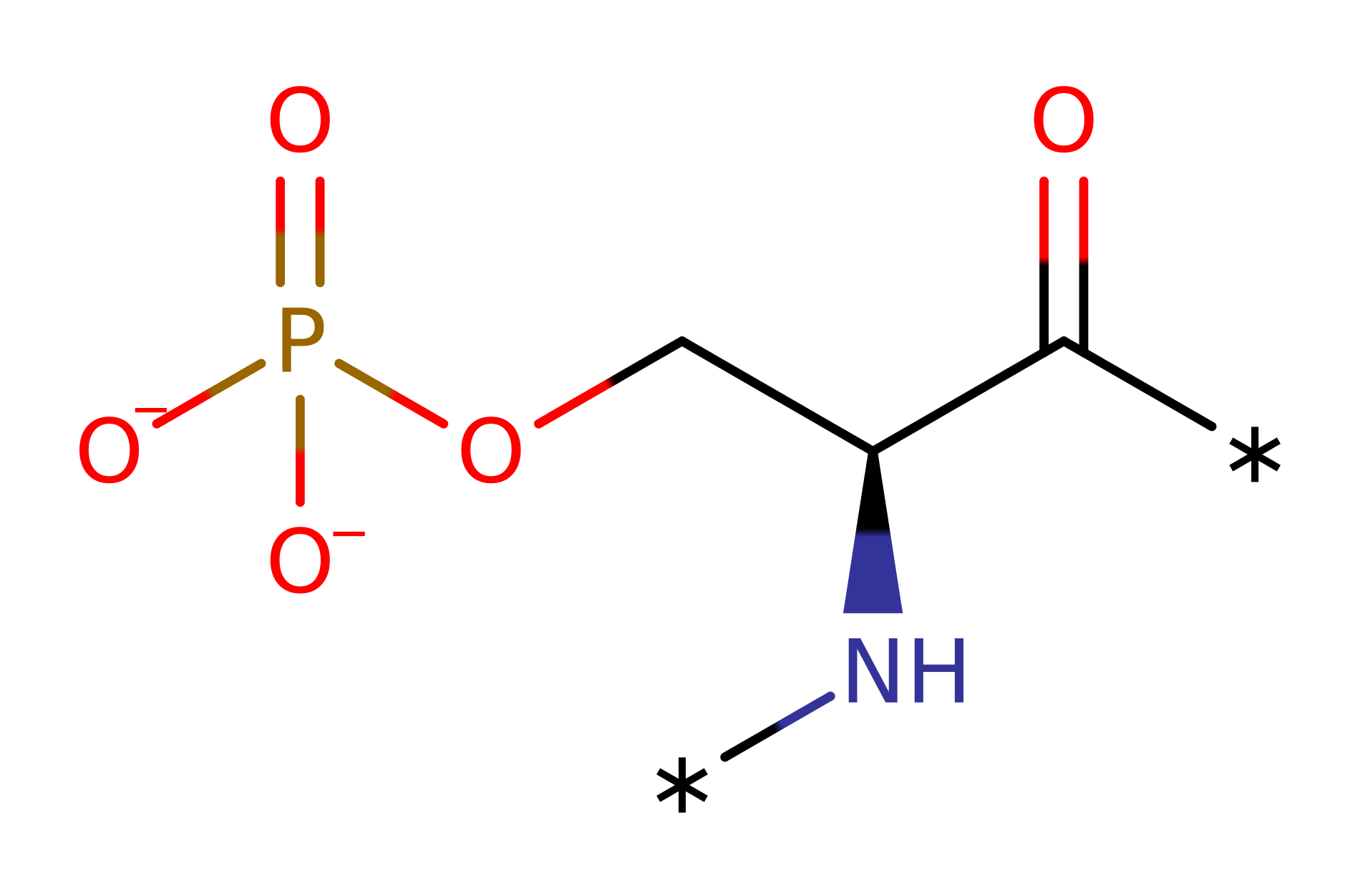
[pyruvate dehydrogenase (acetyl-transferring)] kinase
Mitochondrial Pyruvate Dehydrogenase kinase (PDK) regulates the activity of Pyruvate dehydrogenase complex (PDC) by phosphorylating a serine residue at the E1 subunit and thus inactivating it. Mitochondrial PDK is in a family of serine protein kinases that lacks sequence similarity with all other eukaryotic protein kinase but show similarity with key motifs of prokaryotic histidine protein kinases and a family of eukaryotic ATPases. In mammals, 4 isozymes of PDK have been found, PDK1, PDK2, PDK3, PDK4. PDK2 is found in many tissues but is low in amount in lung and spleen. It only phosphorylates site 2 and 3 serine residue of the E1 subunit.
Reference Protein and Structure
- Sequence
-
Q64536
 (2.7.11.2)
(2.7.11.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rattus norvegicus (Norway rat)

- PDB
-
1jm6
- Pyruvate dehydrogenase kinase, isozyme 2, containing ADP
(2.5 Å)



- Catalytic CATH Domains
-
3.30.565.10
 (see all for 1jm6)
(see all for 1jm6)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.7.11.2)
Enzyme Mechanism
Introduction
One proposed mechanism is a general base mechanism. Glu251 deprotonates the targeted serine residue of the pyruvate dehydrogenase E1 subunit and promotes its nucleophilic attack on the terminal phosphate of ATP. His247 polarises Glu243 to allow deprotonation. Magnesium ion coordinated to Asp helps in positioning of the ATP and promotes the nucleophilic attack. There is also evidence to suggest Glu251 alongside Asn255 and Lys254 in fact coordinates to a magnesium ion while His247 is the base catalyst. Therefore, it is still unclear of the exact mechanism pyruvate dehydrogenase kinase has.
Catalytic Residues Roles
| UniProt | PDB* (1jm6) | ||
| Lys254, Asn255 | Lys1246(254)A, Asn1247(255)A | Proposed to coordinate to a magnesium ion. | metal ligand |
| Glu251 | Glu1243(251)A | A general base to deprotonate the hydroxyl group of the target E1-alpha Serine residue and promotes the nucleophilic attack of the terminal phosphate of ATP or is proposed to coordinate to a magnesium ion. | proton shuttle (general acid/base), metal ligand |
| His247 | His1239(247)A | It is proposed to either polarise Glu251 for deprotonation or act as a general base catalyst. | electrostatic stabiliser |
Chemical Components
References
- Steussy CN et al. (2001), J Biol Chem, 276, 37443-37450. Structure of Pyruvate Dehydrogenase Kinase: NOVEL FOLDING PATTERN FOR A SERINE PROTEIN KINASE. DOI:10.1074/jbc.m104285200. PMID:11483605.
- Tovar-Méndez A et al. (2005), Arch Biochem Biophys, 434, 159-168. Analysis of the catalytic mechanism of pyruvate dehydrogenase kinase. DOI:10.1016/j.abb.2004.10.017. PMID:15629119.
- Tuganova A et al. (2001), J Biol Chem, 276, 17994-17999. An Essential Role of Glu-243 and His-239 in the Phosphotransfer Reaction Catalyzed by Pyruvate Dehydrogenase Kinase. DOI:10.1074/jbc.m009327200. PMID:11278487.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| His1239(247)A | electrostatic stabiliser |
| Glu1243(251)A | proton shuttle (general acid/base) |
| Asn1247(255)A | metal ligand |
| Lys1246(254)A | metal ligand |
| Glu1243(251)A | metal ligand |