DNA nucleotidylexotransferase
Terminal deoxynucleotidyl transferase (TdT) catalyses the condensation of deoxyribonucleotide triphosphates onto the 3' hydroxyl ends of DNA strands in a template-independent manner. It can also catalyse the addition of ribonucleotides and a range of unnatural nucleotides onto DNA strands. TdT has only been found in mammals, where it is highly conserved. It has a role in the generation of combinatorial diversity in lymphocytes, where it functions to add nucleotides (N regions) to the V(D)J recombination junctions of immunoglobulin and T-cell receptor genes. Together with DNA polymerase beta and lambda, TdT belongs to a family of polymerases called pol X, a subclass of an ancient nucleotidyl transferase (NT) superfamily. The active site of this family is structurally similar to that of the pol I and pol alpha families even though the topology of the catalytic domain is different; in all cases the active site is made up of three carboxylate side chains which bind two divalent cations. Tdt is unique in this respect due to its ability to be catalytically active with a range of divalent cations bound, Co2+, Mn2+, Zn2+and Mg2+. The ion bound determines substrate specificity and the enzyme's kinetics.
Reference Protein and Structure
- Sequence
-
P09838
 (2.7.7.31, 3.1.11.-)
(2.7.7.31, 3.1.11.-)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Mus musculus (house mouse)

- PDB
-
1jms
- Crystal Structure of the Catalytic Core of Murine Terminal Deoxynucleotidyl Transferase
(2.36 Å)



- Catalytic CATH Domains
-
3.30.460.10
 (see all for 1jms)
(see all for 1jms)
- Cofactors
- Magnesium(2+) (2)
Enzyme Reaction (EC:2.7.7.31)
Enzyme Mechanism
Introduction
By analogy with DNA polymerase beta, TdT uses an Mg2+ ion to promote attack by the 3' hydroxyl group of the primer on the alpha phosphorous of the incoming NTP which is coupled to the stereochemical inversion. Coordination of the 3' hydroxyl group to the Mg2+ ion lowers the pKa of this group to allow deprotonation by Asp 434 during attack on the NTP. The Mg2+ ion also provides a positive charge to stabilise the pentacoordinate transition state of the alpha phosphorous.
Catalytic Residues Roles
| UniProt | PDB* (1jms) | ||
| Asp343, Asp345 | Asp343(214)A, Asp345(216)A | Interacts with divalent metal ions. | metal ligand |
| Asp434 | Asp434(305)A | Accepts proton from attacking 3' OH group of primer. | metal ligand, proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, overall product formed, overall reactant used, inferred reaction step, native state of enzyme regeneratedReferences
- Delarue M et al. (2002), EMBO J, 21, 427-439. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. DOI:10.1093/emboj/21.3.427. PMID:11823435.
- Beard WA et al. (2014), Biochemistry, 53, 2768-2780. Structure and mechanism of DNA polymerase β. DOI:10.1021/bi500139h. PMID:24717170.
- Bebenek K et al. (2014), Biochemistry, 53, 2781-2792. Structure-function studies of DNA polymerase λ. DOI:10.1021/bi4017236. PMID:24716527.
- Motea EA et al. (2010), Biochim Biophys Acta, 1804, 1151-1166. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. DOI:10.1016/j.bbapap.2009.06.030. PMID:19596089.
- Cisneros GA et al. (2008), DNA Repair (Amst), 7, 1824-1834. Catalytic mechanism of human DNA polymerase lambda with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies. DOI:10.1016/j.dnarep.2008.07.007. PMID:18692600.
- Boulé JB et al. (2001), J Biol Chem, 276, 31388-31393. Terminal Deoxynucleotidyl Transferase Indiscriminately Incorporates Ribonucleotides and Deoxyribonucleotides. DOI:10.1074/jbc.m105272200. PMID:11406636.
- Pelletier H et al. (1994), Science, 264, 1891-1903. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. DOI:10.2210/pdb2bpg/pdb. PMID:7516580.
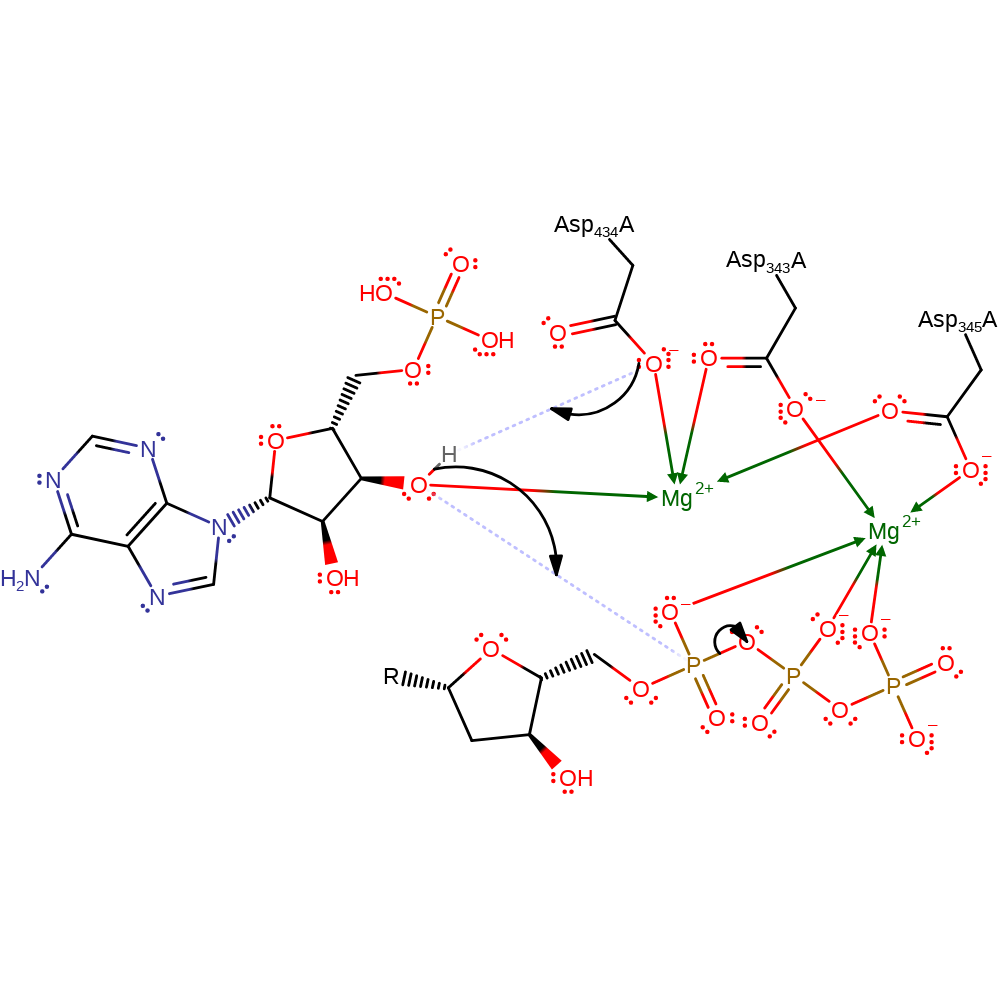
Step 1. Mg2+A lowers the pKa around the 3'OH on RNA, facilitating protonation of Asp434. This inevitably increases the 3'OH nucleophilicity to attack the alpha phosphate on the incoming DTP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp343(214)A | metal ligand |
| Asp345(216)A | metal ligand |
| Asp434(305)A | metal ligand |
| Asp434(305)A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, overall product formed, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp434(305)A | proton donor |


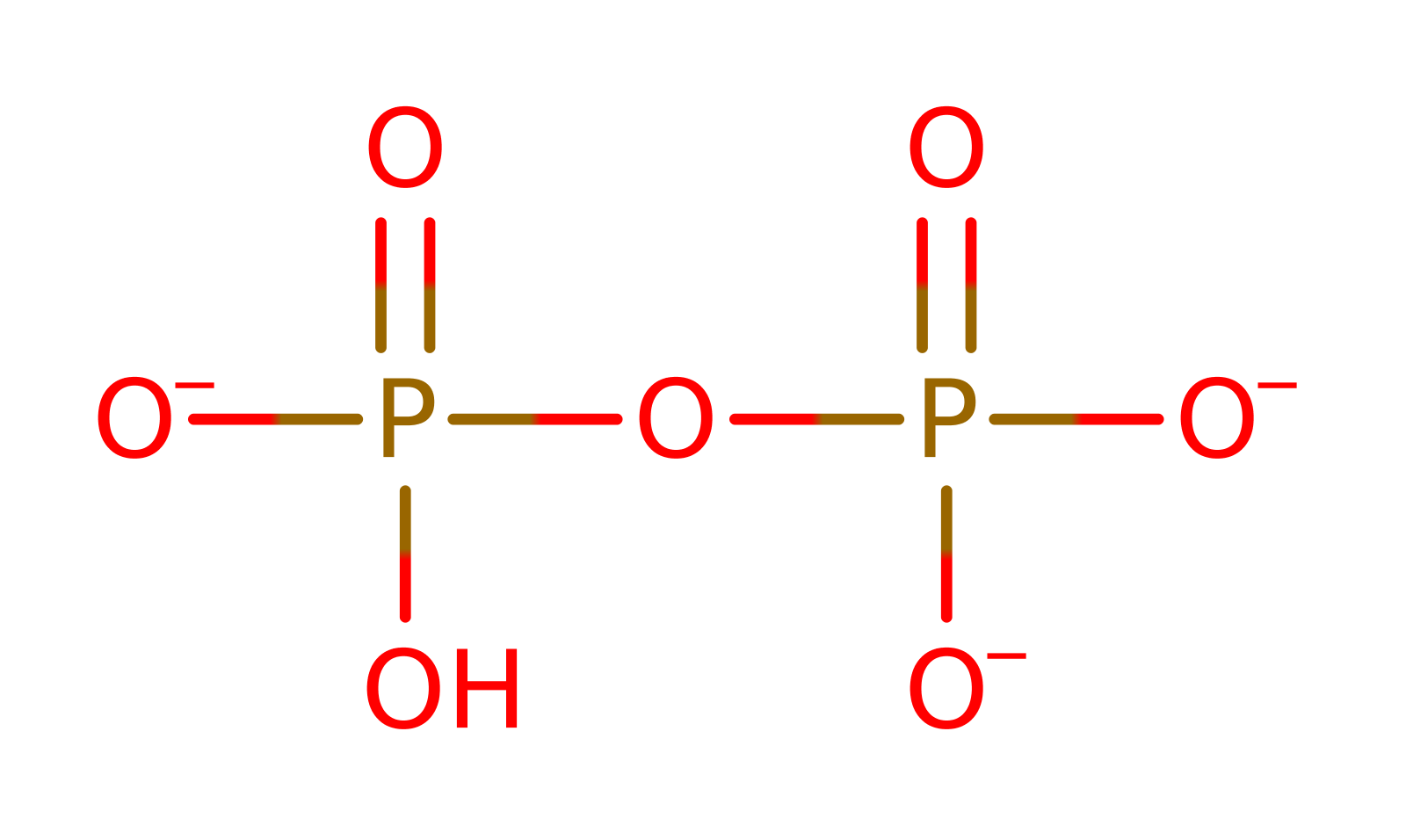

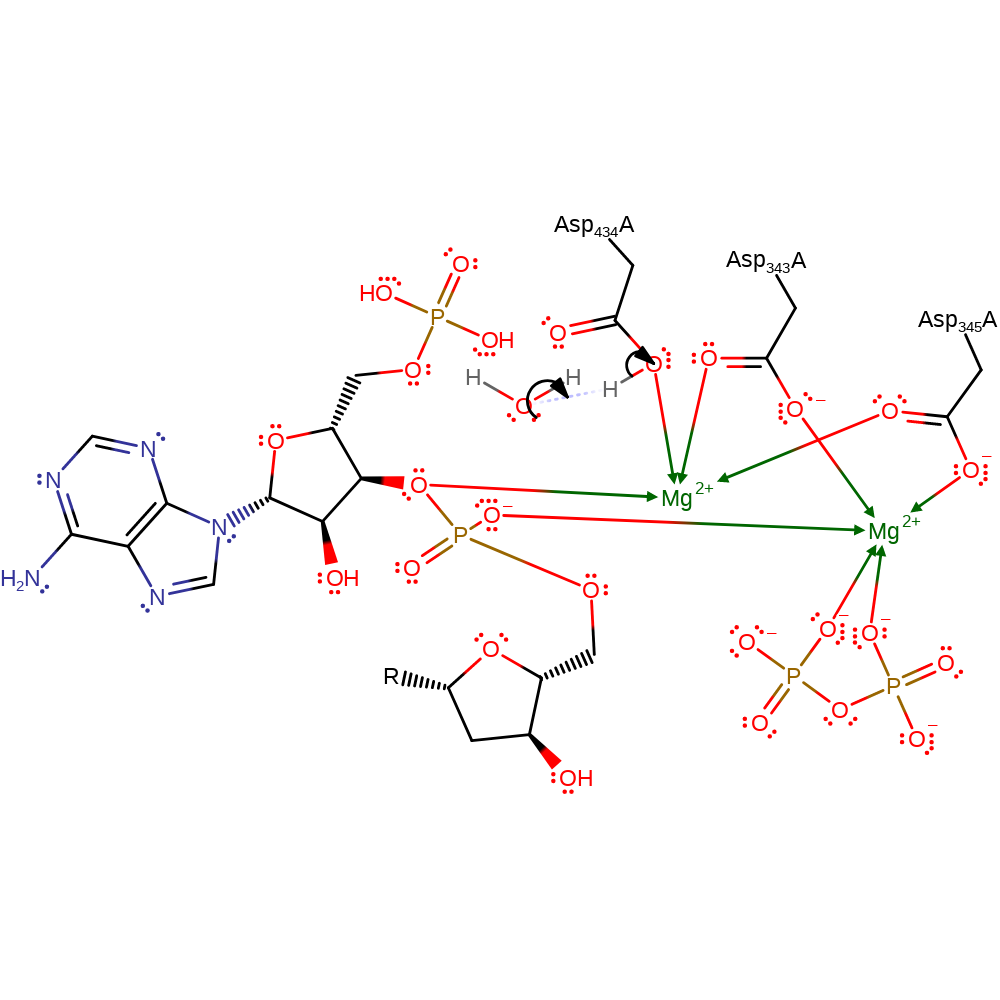
 Download:
Download: