Acetate kinase
Acetate kinase is a key enzyme for many methanogenic bacteria as it is able to catalyse the interconversion of acetate and acetyl phosphate using the removal of a phosphate from ATP. Structural analysis of the enzyme from Methanosarcina thermophila, an Archaea, shows that it is a member of the sugar kinase superfamily which includes hexokinase, and geochemical studies suggest that it is most likely to be the urkinase- the ancestor of all of the members of the superfamily.
Reference Protein and Structure
- Sequence
-
P38502
 (2.7.2.1)
(2.7.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Methanosarcina thermophila (Archaea)

- PDB
-
1g99
- AN ANCIENT ENZYME: ACETATE KINASE FROM METHANOSARCINA THERMOPHILA
(2.5 Å)



- Catalytic CATH Domains
-
3.30.420.40
 (see all for 1g99)
(see all for 1g99)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:2.7.2.1)
Enzyme Mechanism
Introduction
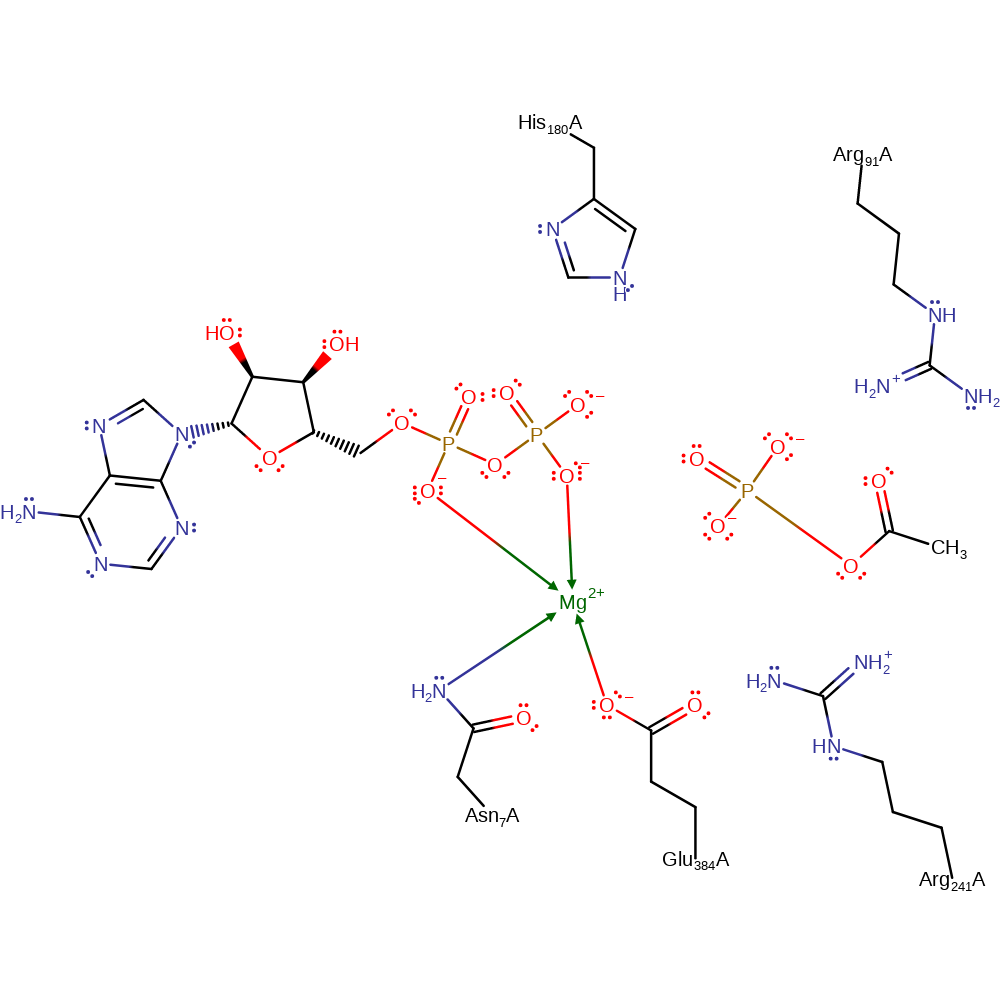
The proposed mechanism is an in-line single displacement reaction where the acetate acts as the nucleophile to attack the gamma phosphate of ATP, resulting in acetyl phosphate and ADP. For this reaction to take place, a magnesium ion, His180, Arg241 and Arg91 are important in stabilising the reaction transition state.
Catalytic Residues Roles
| UniProt | PDB* (1g99) | ||
| Asn7 | Asn7A | Coordinates to the magnesium ion which in turn stabilises the transition state in the reaction. | metal ligand |
| Glu384 | Glu384A | Coordinates to the magnesium ion which in turn helps stabilise the transition state in the reaction. | metal ligand |
| His180 | His180A | Coordinates to the equatorial oxygens on ATP to help stabilise the transition state. | electrostatic stabiliser, polar interaction |
| Arg91 | Arg91A | Contacts alpha phosphate of ATP thus stabilises the phosphate transition state through electrostatic interactions. It also has been proposed to help position acetate for nucleophilic attack in the reaction. | electrostatic stabiliser, polar interaction |
| Arg241 | Arg241A | Contacts alpha phosphate of ATP helping stabilise the transition state. | electrostatic stabiliser, polar interaction |
Chemical Components
bimolecular nucleophilic substitution, overall product formed, overall reactant usedReferences
- Gorrell A et al. (2005), J Biol Chem, 280, 10731-10742. Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila. DOI:10.1074/jbc.M412118200. PMID:15647264.
- Buss KA et al. (2001), J Bacteriol, 183, 680-686. Urkinase: Structure of Acetate Kinase, a Member of the ASKHA Superfamily of Phosphotransferases. DOI:10.1128/jb.183.2.680-686.2001. PMID:11133963.
- Miles RD et al. (2001), J Biol Chem, 276, 45059-45064. Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina thermophila. DOI:10.1074/jbc.m108355200. PMID:11562377.

Step 1. Acetate's carboxyl group acts as a nucleophile and attacks and displaces the gamma-phosphate on ATP via a trigonal bypyramidal transition state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg91A | electrostatic stabiliser |
| Arg241A | electrostatic stabiliser |
| Asn7A | metal ligand |
| Glu384A | metal ligand |
| His180A | electrostatic stabiliser |
| Arg91A | polar interaction |
| His180A | polar interaction |
| Arg241A | polar interaction |




 Download:
Download: