Adenylosuccinate synthase
Adenylosuccinate synthetase is an essential enzyme in Escherichia coli and many other forms of life, catalysing the first committed step in de novo biosyntheses of AMP: The catalytic mechanism proceeds with the initial formation of 6-phosphoryl-IMP by nucleophilic attack of the 6-oxo group of IMP on the gamma-phosphorus atom of GTP, followed by the nucleophilic displacement of the 6-phosphoryl group by L -aspartate to form adenylosuccinate.
Reference Protein and Structure
- Sequence
-
P0A7D4
 (6.3.4.4)
(6.3.4.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1gim
- CRYSTAL STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH GDP, IMP, HADACIDIN, NO3-, AND MG2+. DATA COLLECTED AT 100K (PH 6.5)
(2.5 Å)



- Catalytic CATH Domains
-
3.40.440.10
 (see all for 1gim)
(see all for 1gim)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.3.4.4)
Enzyme Mechanism
Introduction
There is a two-step nature of the reaction. Four residues are essential for the reaction: Asp13, His41 and Gln224 and Lys16 . Asp13 acts first as a catalytic base, removing a proton from the N1 atom of IMP, while His41 acts as an acid, protonating the beta-phosphoryl group of GTP which then becomes the leaving group. It is suggested that once the IMP-6-phosphoryl intermediate is formed, as a response to binding aspartate the active site alters: Asp13 moves to be co-ordinated by the Mg2+ ion, transforming it into an acid, while His41 rotates to interact with the 6-phosphoryl group instead of the GDP phosphate. Both of these changes facilitate the displacement of phosphate by aspartate. Throughout the reaction Gln224 is positioned to H-bond to the 6-0 atom of IMP, stabilising the negative charge that develops on it. Similarly Lys16 hydrogen bonds with the 6-phosphoryl group of the intermediate.
Catalytic Residues Roles
| UniProt | PDB* (1gim) | ||
| Lys17 | Lys16A | Lys16 hydrogen bonds with the 6-phosphoryl group of the intermediate to stabilise it and also, perhaps with beta-phosphoryl group of GDP. | hydrogen bond donor, electrostatic stabiliser |
| Gln225 | Gln224A | Gln224 works in concert with asp13 to stabilise the 6-oxyanion of IMP via hydrogen bonding. | hydrogen bond donor, electrostatic stabiliser |
| Asp14, Gly41 (main-C) | Asp13A, Gly40A (main-C) | Forms part of the magnesium binding site. | hydrogen bond acceptor, hydrogen bond donor, metal ligand, proton acceptor, proton donor, electrostatic stabiliser |
| His42 | His41A | His41 acts as a catalytic acid in the phosphorylation, by proton donation to the leaving group GDP via binding to beta-phosphoryl group of GTP. Then again as an acid in the phosphoryl displacement by donating a proton to the phosphate group. The conformation change during the second step is thought to be triggered by the association of L-aspartate. Note it also acts throughout the reaction to stabilise the intermediate, as the NE2 of His41 hydrogen bonds with the 6-phosphoryl group of the intermediate. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol), overall reactant used, intermediate formation, bimolecular nucleophilic substitution, overall product formed, aromatic bimolecular nucleophilic substitution, intermediate collapse, intermediate terminated, dephosphorylation, proton relay, native state of enzyme regenerated, inferred reaction stepReferences
- Choe JY et al. (1999), Biochemistry, 38, 6953-6961. Mechanistic Implications from Crystalline Complexes of Wild-Type and Mutant Adenylosuccinate Synthetases fromEscherichia coli†,‡. DOI:10.1021/bi990159s. PMID:10346917.
- Iancu CV et al. (2006), Biochemistry, 45, 11703-11711. Cavitation as a Mechanism of Substrate Discrimination by Adenylosuccinate Synthetases†,‡. DOI:10.1021/bi0607498. PMID:16981730.
- Gorrell A et al. (2002), J Biol Chem, 277, 8817-8821. Determinants of L-Aspartate and IMP Recognition inEscherichia coli Adenylosuccinate Synthetase. DOI:10.1074/jbc.m111810200. PMID:11781326.
- Hou Z et al. (2002), J Biol Chem, 277, 5970-5976. IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli. DOI:10.1074/jbc.m109561200. PMID:11741996.
- Honzatko RB et al. (1999), Arch Biochem Biophys, 370, 1-8. Structure–Function Studies of Adenylosuccinate Synthetase from Escherichia coli. DOI:10.1006/abbi.1999.1383. PMID:10496970.
- Kang C et al. (1997), J Biol Chem, 272, 11881-11885. Residues Essential for Catalysis and Stability of the Active Site of Escherichia coli Adenylosuccinate Synthetase as Revealed by Directed Mutation and Kinetics. DOI:10.1074/jbc.272.18.11881. PMID:9115248.
- Wang W et al. (1997), J Biol Chem, 272, 16911-16916. Relationship of Conserved Residues in the IMP Binding Site to Substrate Recognition and Catalysis in Escherichia coliAdenylosuccinate Synthetase. DOI:10.1074/jbc.272.27.16911. PMID:9202000.
- Poland BW et al. (1997), J Biol Chem, 272, 15200-15205. Entrapment of 6-Thiophosphoryl-IMP in the Active Site of Crystalline Adenylosuccinate Synthetase from Escherichia coli. DOI:10.1074/jbc.272.24.15200. PMID:9182542.
- Liu F et al. (1992), J Biol Chem, 267, 2388-2392. Site-directed mutagenesis of the phosphate-binding consensus sequence in Escherichia coli adenylosuccinate synthetase. PMID:1733940.

Step 1. Asp13 deprotonates IMP with concomitant tautomerisation to give the oxyanion, which is stabilised by a hydrogen bond from Gln224. Activated IMP is produced. His41 and Mg stabilise the GTP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp13A | hydrogen bond acceptor |
| His41A | hydrogen bond donor, electrostatic stabiliser |
| Gln224A | hydrogen bond donor, electrostatic stabiliser |
| Lys16A | hydrogen bond donor, electrostatic stabiliser |
| Asp13A | metal ligand |
| Gly40A (main-C) | metal ligand |
| Asp13A | proton acceptor |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol), overall reactant used, intermediate formation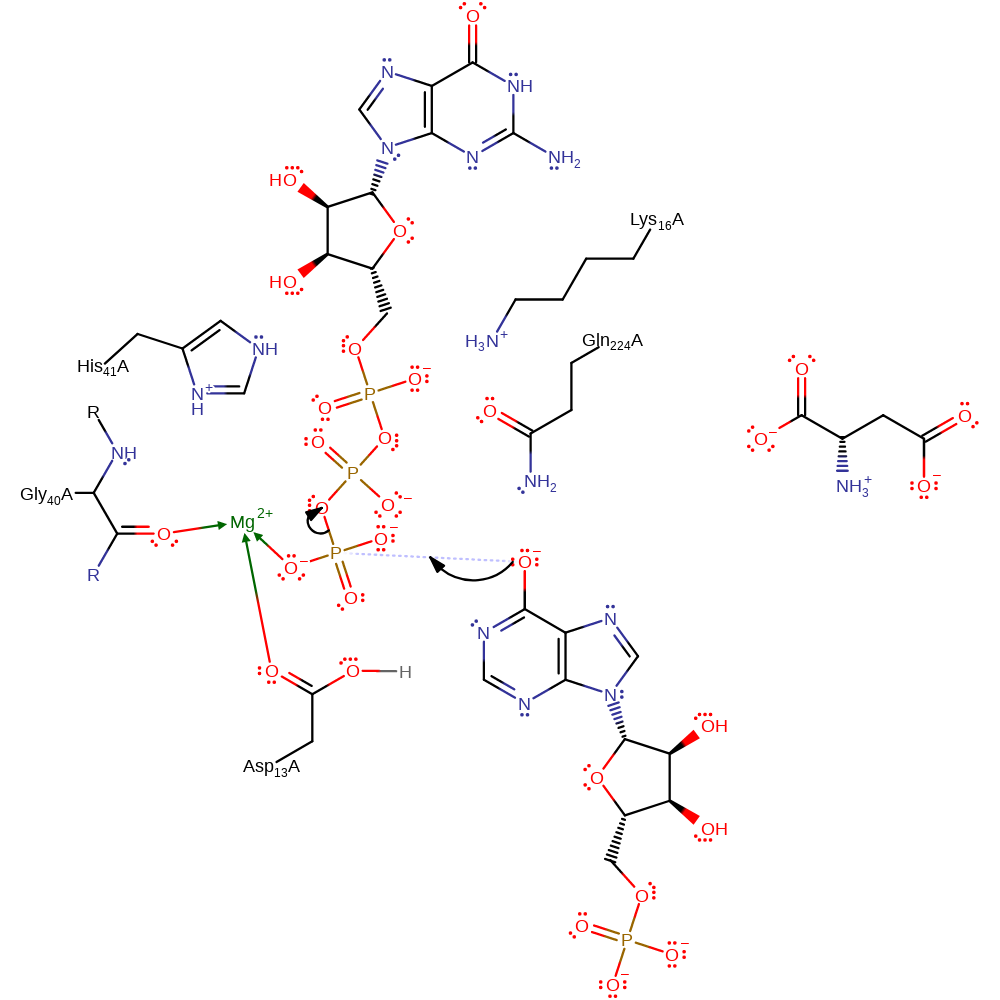
Step 2. Activated IMP acts as a nucleophile and attacks the gamma-phosphate of GTP in a substitution reaction. GDP and phosphorylated IMP are produced. His41 and Mg stabilise the GTP, Asp13 and Gln223 stabilise the activated IMP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp13A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| His41A | electrostatic stabiliser, hydrogen bond donor |
| Gln224A | hydrogen bond donor, electrostatic stabiliser |
| Lys16A | hydrogen bond donor, electrostatic stabiliser |
| Asp13A | metal ligand |
| Gly40A (main-C) | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation
Step 3. The amine nitrogen of L-aspartate acts as a nucleophile upon the C6 of the phosphorylated IMP in a substitution reaction at an aromatic species. The IMP intermediate is dephosphorylated. The liberated phosphate group accepts a proton from the stabilising His41. Asp13 and Gln223 continue to stabilise the IMP intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp13A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| His41A | electrostatic stabiliser, hydrogen bond donor |
| Gln224A | hydrogen bond donor, electrostatic stabiliser |
| Lys16A | hydrogen bond donor |
| Asp13A | metal ligand |
| Gly40A (main-C) | metal ligand |
| His41A | proton donor |
Chemical Components
ingold: aromatic bimolecular nucleophilic substitution, proton transfer, overall reactant used, overall product formed, intermediate collapse, intermediate terminated, dephosphorylation
Step 4. Inferred return step in which His41 deprotonates the bound phosphate, which in turn deprotonates Asp13. It is assumed that the positive nitrogen of the product loses its extra proton to either one of the carboxylate groups (shown) or bulk solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp13A | hydrogen bond donor |
| His41A | hydrogen bond acceptor |
| Asp13A | metal ligand |
| Gly40A (main-C) | metal ligand |
| Asp13A | proton donor |
| His41A | proton acceptor |




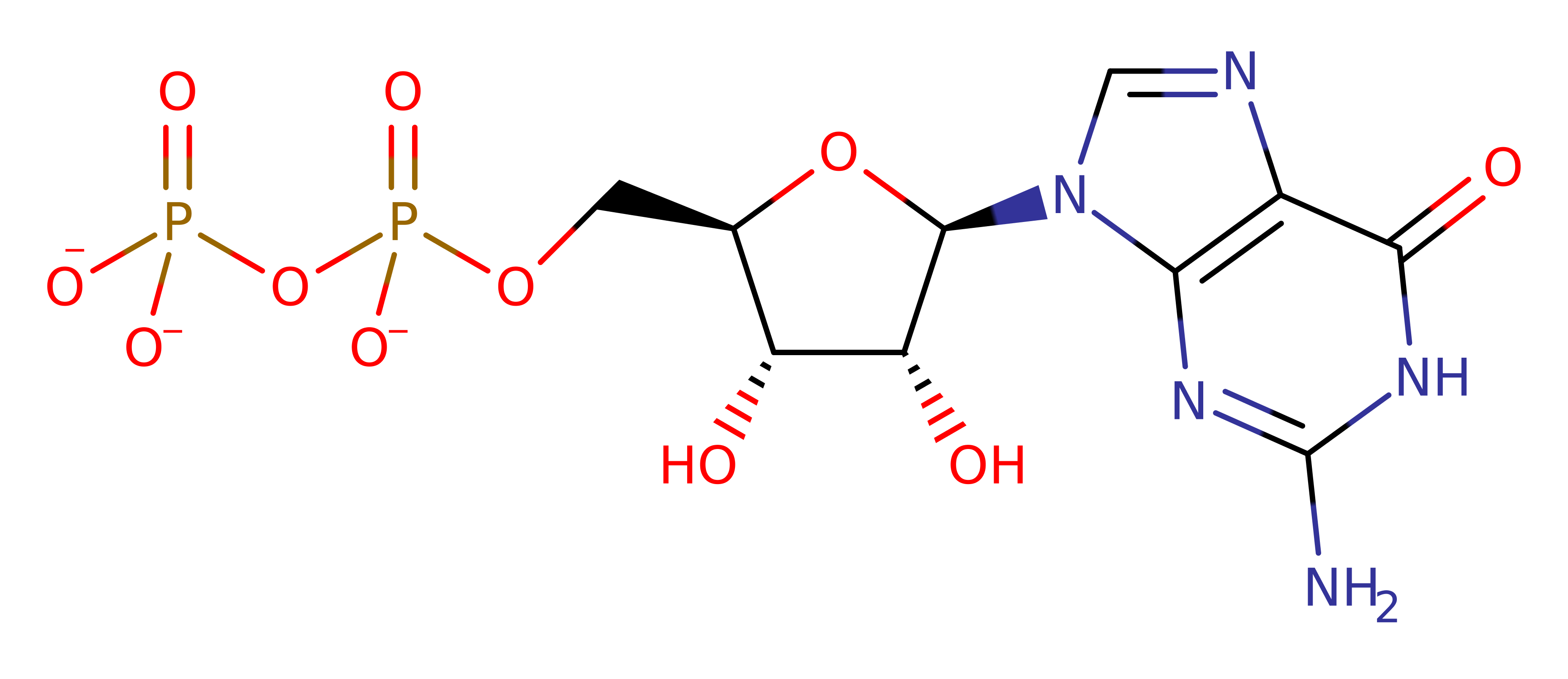


 Download:
Download: