4-hydroxy-2-oxovalerate aldolase
4-hydroxy-2-ketovalerate aldolase catalyses the conversion of 4-hydroxy-2-ketovalerate to acetaldehyde and pyruvate. It associates tightly with acetaldehyde dehydrogenase and appears to be inactive when expressed without this dehydrogenase. The aldolase and dehydrogenase form a bifunctional enzyme, and there is evidence that the reactive acetaldehyde intermediate is passed between the two active sites via a channelling tunnel. the bifunctional enzyme catalyses the final two steps in the degradation of catechol, an intermediate in the degradation of aromatic compounds by many bacteria.
Reference Protein and Structure
- Sequences
-
P51016
 (4.1.3.39)
(4.1.3.39)
Q52060 (1.2.1.10)
(1.2.1.10)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas sp. CF600 (Bacteria)

- PDB
-
1nvm
- Crystal structure of a bifunctional aldolase-dehydrogenase : sequestering a reactive and volatile intermediate
(1.7 Å)



- Catalytic CATH Domains
-
3.20.20.70
 1.10.8.60
1.10.8.60  (see all for 1nvm)
(see all for 1nvm)
- Cofactors
- Water (1), Manganese(2+) (1)
Enzyme Reaction (EC:4.1.3.39)
Enzyme Mechanism
Introduction
The proposed catalytic mechanism involves His 21 acting as a base to remove a proton from the 4-hydroxyl group. Concomitant cleavage of the C3-C4 bond leads to formation of acetaldehyde and the enolate of pyruvate. Accumulation of negative charge on the 2-carbonyl oxygen during formation of the enolate is stabilised by an Mn2+ ion. The enolate is protonated on C3 by Tyr 291 (which is later re-protonated by His 21) to form pyruvate, while the acetaldehyde is passed to the dehydrogenase active site via a tunnel. Access to this tunnel is proposed to be controlled by movements of Tyr 291 during the catalytic cycle.
Catalytic Residues Roles
| UniProt | PDB* (1nvm) | ||
| Asp18, His200, His202 | Asp18A, His200A, His202A | Coordinate the metal ligand. | metal ligand |
| His21 | His21A | Deprotonates the 4-hydroxyl group of the substrate, leading to generation of acetalydehyde and the enolate of pyruvate. Later protonates Tyr 291. | proton acceptor, proton donor |
| Tyr291 | Tyr291A | Protonates the methylene group of the enolate of pyruvate. Regains a proton from His 21. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular elimination, overall reactant used, overall product formed, assisted keto-enol tautomerisation, native state of enzyme regeneratedReferences
- Manjasetty BA et al. (2003), Proc Natl Acad Sci U S A, 100, 6992-6997. Crystal structure of a bifunctional aldolase-dehydrogenase: Sequestering a reactive and volatile intermediate. DOI:10.1073/pnas.1236794100. PMID:12764229.

Step 1. His 21 acts as a base and deprotonates the C4 hydroxyl group. The concomitant bond cleavage of the C3-C4 bond leads to the formation of acetaldehyde and the enolate of pyruvate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp18A | metal ligand |
| His200A | metal ligand |
| His202A | metal ligand |
| His21A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular elimination, overall reactant used, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp18A | metal ligand |
| His200A | metal ligand |
| His202A | metal ligand |
| Tyr291A | proton acceptor |
| His21A | proton donor |
Chemical Components
proton transfer
Step 3. The enolate is then protonated by Tyr291 and pyruvate is released.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp18A | metal ligand |
| His200A | metal ligand |
| His202A | metal ligand |
| Tyr291A | proton donor |


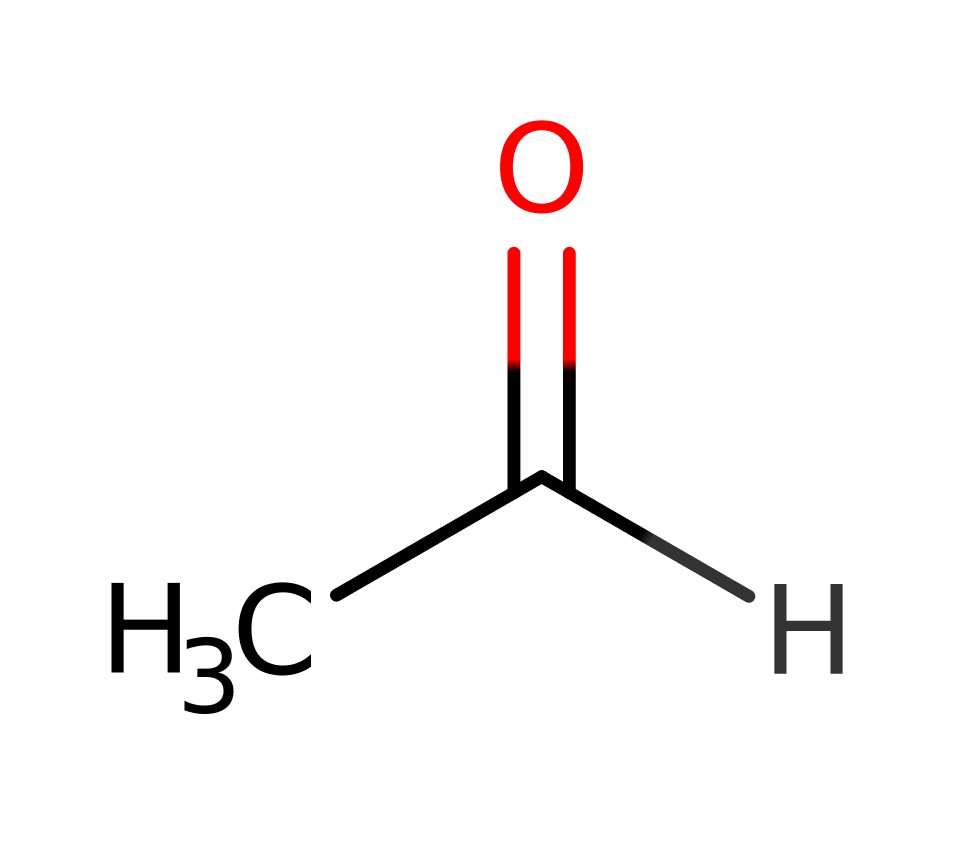

 Download:
Download: