Isovaleryl-CoA dehydrogenase
Isovaryl-CoA dehydrogenase (IVD) belongs to an important flavoprotein family of acyl CoA dehydrogenases that catalyse the alpha,beta-dehydrogenation of their various thioester substrates. IVD is most reactive for a small, branched-chain substrate, catalysing the conversion of isovaleryl-CoA (3-methylbutyryl-CoA) into 3-methylcrotonyl-CoA as the third step of leucine catabolism. This enzyme is a tetrameric flavoprotein which contains one molecule of flavin adenine dinucleotide (FAD) per monomer. Deficiency of IVD results in isovaleric acidemia (IVA), a life-threatening disorder in which leucine catabolism is blocked and organic acids accumulate in the body.
Reference Protein and Structure
- Sequence
-
P26440
 (1.3.8.1, 1.3.8.4)
(1.3.8.1, 1.3.8.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1ivh
- STRUCTURE OF HUMAN ISOVALERYL-COA DEHYDROGENASE AT 2.6 ANGSTROMS RESOLUTION: STRUCTURAL BASIS FOR SUBSTRATE SPECIFICITY
(2.6 Å)



- Catalytic CATH Domains
-
1.20.140.10
 2.40.110.10
2.40.110.10  (see all for 1ivh)
(see all for 1ivh)
- Cofactors
- Fadh2(2-) (1)
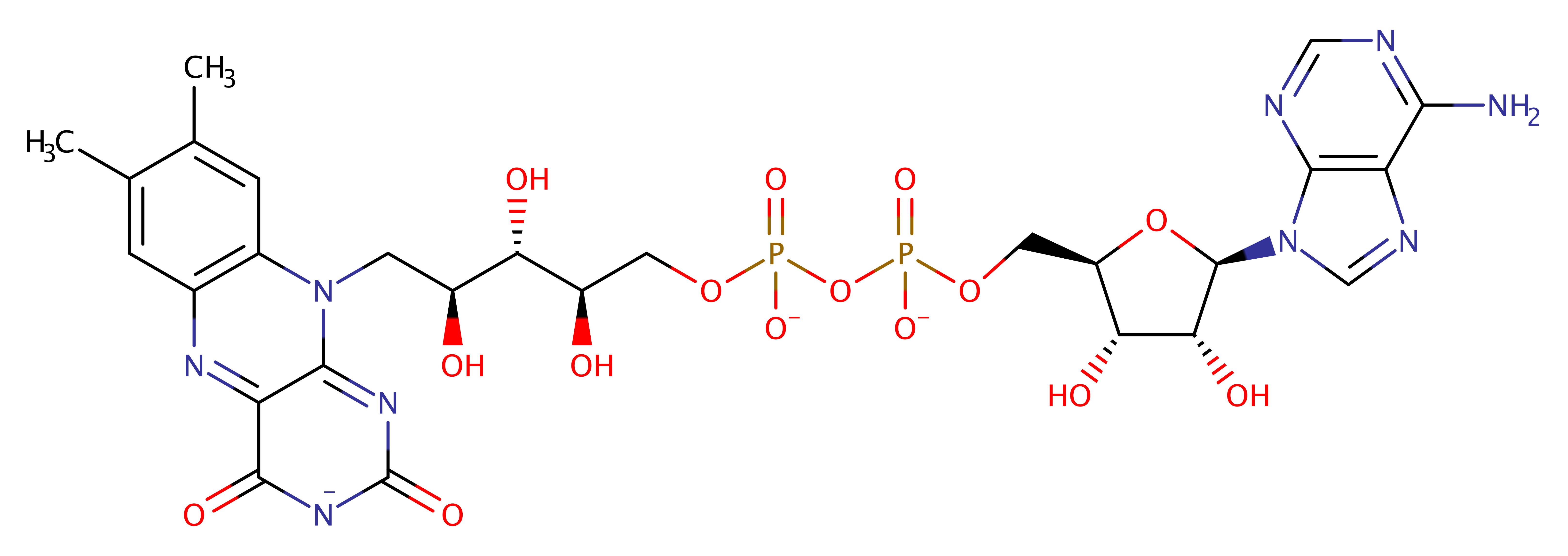
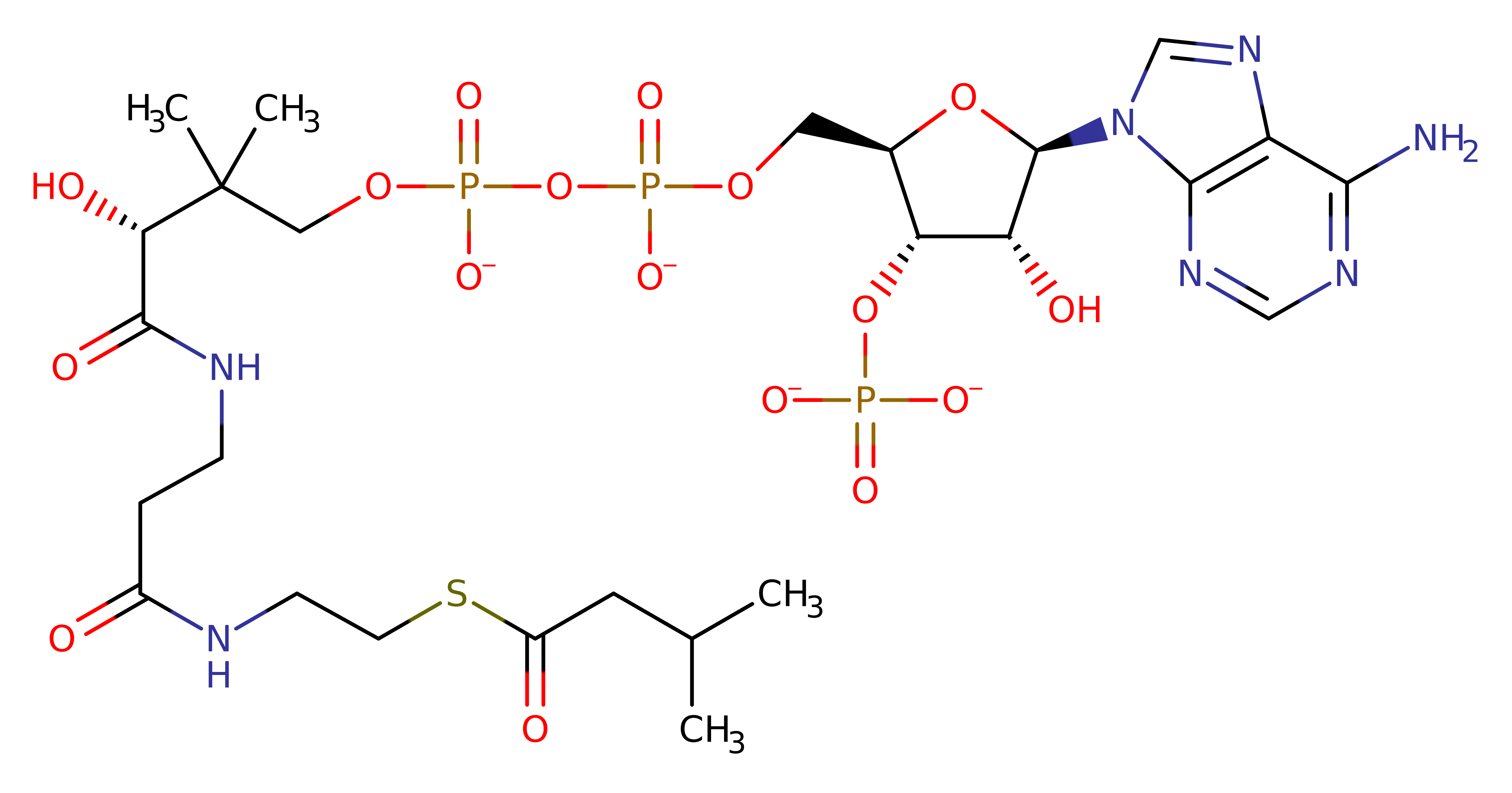
Enzyme Reaction (EC:1.3.8.4)
Enzyme Mechanism
Introduction
The dehydrogenation reaction is initiated as Glu254 acts as a catalytic base abstracting the R -hydrogen of substrate in the form of a proton, with a concomitant transfer of the alpha -hydrogen from substrate as a hydride to the N(5) position of the isoalloxazine ring of the FAD, forming a trans double bond between the C(alpha) and C(beta) of the fatty acyl-CoA substrate. Ultimately, reducing equivalents are passed to the mitochondrial respiratory chain first
via electron transfer flavoprotein (ETF) the physiological electron acceptor for all the acyl-CoA dehydrogenases, and second by ETF- ubiquinone oxidoreductase, a membrane-bound Fe-S flavoprotein.
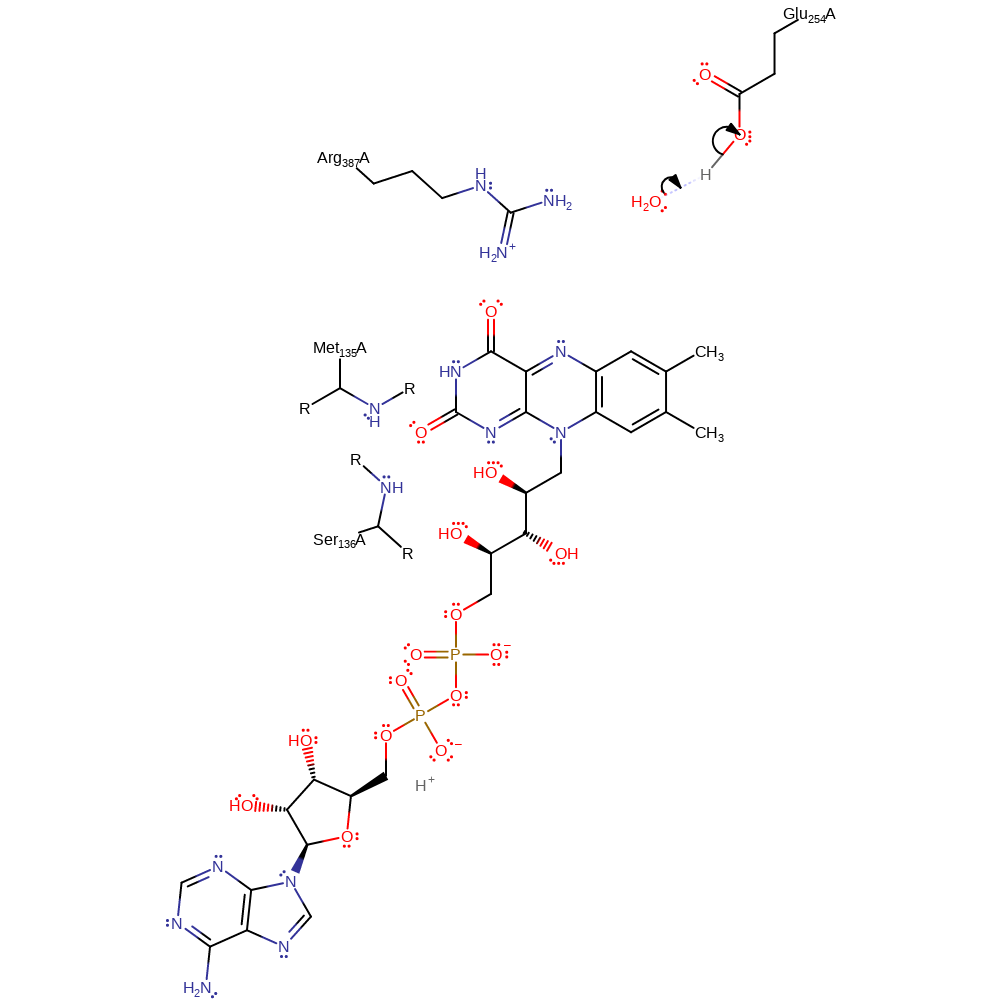
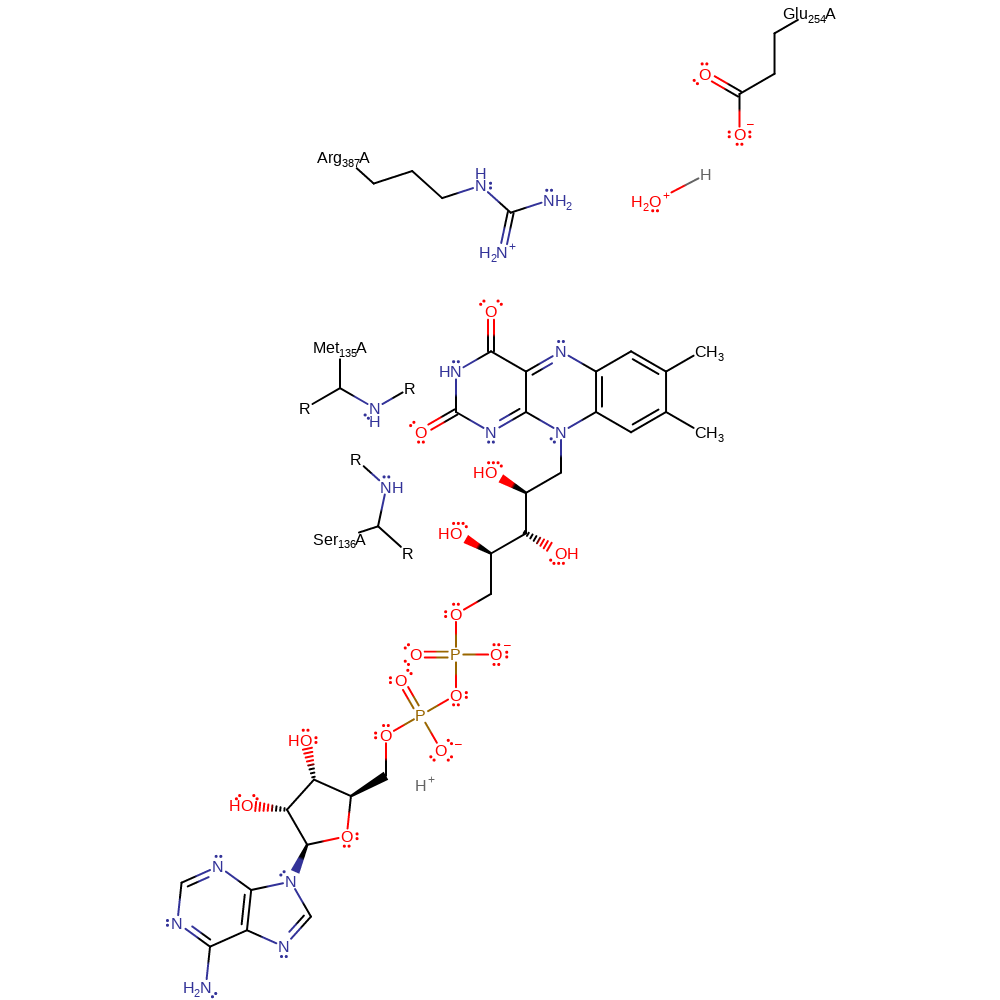
Note residues Ser136 and Met135 may serve to stabilise the oxyanion hole created in reduced FAD, however their role has not been specifically proven by experimental evidence.
Catalytic Residues Roles
| UniProt | PDB* (1ivh) | ||
| Arg416 | Arg387A | Involved in binding and stabilising the enzyme-substrate complex. | electrostatic stabiliser |
| Glu283 | Glu254A | Acts as a general base to abstract a proton from C2 of isovaleryl-CoA, in the dehydrogenation process. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, steric role |
| Ser165 (main-N), Met164 (main-N) | Ser136A (main-N), Met135A (main-N) | Forms part of the oxyanion hole. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular elimination, hydride transfer, aromatic bimolecular nucleophilic addition, overall reactant used, intermediate formation, cofactor used, overall product formed, electron transfer, radical formation, radical termination, proton transfer, intermediate terminated, native state of cofactor regenerated, native state of enzyme regenerated, inferred reaction stepReferences
- Ghisla S et al. (2004), Eur J Biochem, 271, 494-508. Acyl-CoA dehydrogenases. A mechanistic overview. DOI:10.1046/j.1432-1033.2003.03946.x. PMID:14728676.
- Mohsen AW et al. (2015), Biochimie, 108, 108-119. Kinetic and spectral properties of isovaleryl-CoA dehydrogenase and interaction with ligands. DOI:10.1016/j.biochi.2014.11.007. PMID:25450250.
- Ozgul RK et al. (2014), Eur J Med Genet, 57, 596-601. Phenotypic and genotypic spectrum of Turkish patients with isovaleric acidemia. DOI:10.1016/j.ejmg.2014.08.006. PMID:25220015.
- Bei F et al. (2013), Gene, 524, 396-400. Two novel isovaleryl-CoA dehydrogenase gene mutations in a Chinese infant. DOI:10.1016/j.gene.2013.03.139. PMID:23587913.
- Dercksen M et al. (2012), J Inherit Metab Dis, 35, 1021-1029. Clinical variability of isovaleric acidemia in a genetically homogeneous population. DOI:10.1007/s10545-012-9457-2. PMID:22350545.
- Vatanavicharn N et al. (2011), Pediatr Int, 53, 990-994. Phenotypic and mutation spectrums of Thai patients with isovaleric acidemia. DOI:10.1111/j.1442-200x.2011.03488.x. PMID:22004070.
- Lin WD et al. (2007), Mol Genet Metab, 90, 134-139. Genetic mutation profile of isovaleric acidemia patients in Taiwan. DOI:10.1016/j.ymgme.2006.08.011. PMID:17027310.
- Lee YW et al. (2007), Mol Genet Metab, 92, 71-77. Different spectrum of mutations of isovaleryl-CoA dehydrogenase (IVD) gene in Korean patients with isovaleric acidemia. DOI:10.1016/j.ymgme.2007.05.003. PMID:17576084.
- Goetzman ES et al. (2006), Mol Genet Metab, 87, 233-242. Functional analysis of acyl-CoA dehydrogenase catalytic residue mutants using surface plasmon resonance and circular dichroism. DOI:10.1016/j.ymgme.2005.09.027. PMID:16376132.
- Goetzman ES et al. (2005), J Biol Chem, 280, 4873-4879. Convergent Evolution of a 2-Methylbutyryl-CoA Dehydrogenase from Isovaleryl-CoA Dehydrogenase inSolanum tuberosum. DOI:10.1074/jbc.m412640200. PMID:15574432.
- Nasser I et al. (2004), Biochim Biophys Acta, 1690, 22-32. Thermal unfolding of medium-chain acyl-CoA dehydrogenase and iso(3)valeryl-CoA dehydrogenase: study of the effect of genetic defects on enzyme stability. DOI:10.1016/j.bbadis.2004.04.008. PMID:15337167.
- Volchenboum SL et al. (2001), Mol Genet Metab, 74, 226-237. Arginine 387 of Human Isovaleryl-CoA Dehydrogenase Plays a Crucial Role in Substrate/Product Binding. DOI:10.1006/mgme.2001.3234. PMID:11592819.
- Mohsen AW et al. (1998), Biochemistry, 37, 10325-10335. Characterization of Molecular Defects in Isovaleryl-CoA Dehydrogenase in Patients with Isovaleric Acidemia†. DOI:10.1021/bi973096r. PMID:9665741.
- Tiffany KA et al. (1997), Biochemistry, 36, 8455-8464. Structure of Human Isovaleryl-CoA Dehydrogenase at 2.6 Å Resolution: Structural Basis for Substrate Specificity†,‖. DOI:10.1021/bi970422u. PMID:9214289.
- Schaller RA et al. (1997), Biochemistry, 36, 7761-7768. Mechanism-Based Inhibitor Discrimination in the Acyl-CoA Dehydrogenases†. DOI:10.1021/bi970095q. PMID:9201918.
- Thorpe C et al. (1995), FASEB J, 9, 718-725. Structure and mechanism of action of the acyl-CoA dehydrogenases. PMID:7601336.
- Mohsen AW et al. (1995), Biochemistry, 34, 10146-10152. Identification of the active site catalytic residue in human isovaleryl-CoA dehydrogenase. PMID:7640268.
- Vockley J et al. (1991), Am J Hum Genet, 49, 147-157. Molecular characterization of four different classes of mutations in the isovaleryl-CoA dehydrogenase gene responsible for isovaleric acidemia. PMID:2063866.
- Finocchiaro G et al. (1987), J Biol Chem, 262, 7982-7989. Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases from human liver. PMID:3597357.

Step 1. Glu254 deprotonates C2 of the 3-methylbutanoyl-CoA which initiates the elimination of a hydride ion from the C3, forming 3-methylbut-2-enoyl-CoA and reduced FAD.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met135A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser136A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Glu254A | hydrogen bond acceptor, hydrogen bond donor, steric role |
| Arg387A | electrostatic stabiliser |
| Glu254A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, intermediate formation, cofactor used, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met135A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser136A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Glu254A | hydrogen bond donor |
| Arg387A | electrostatic stabiliser |
Chemical Components
electron transfer, radical formation, overall reactant used, intermediate formation, overall product formed
Step 3. The second single electron transfer from FAD to ETF, which is facilitated by the abstraction of a proton from the FAD by an unidentified base (shown here as a hydroxide ion).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met135A (main-N) | hydrogen bond donor |
| Ser136A (main-N) | hydrogen bond donor |
| Glu254A | hydrogen bond donor |
| Arg387A | electrostatic stabiliser |
Chemical Components
electron transfer, radical termination, proton transfer, overall reactant used, intermediate terminated, native state of cofactor regenerated, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met135A (main-N) | hydrogen bond donor |
| Ser136A (main-N) | hydrogen bond donor |
| Glu254A | hydrogen bond donor |
| Glu254A | proton donor |




 Download:
Download: