Purine-nucleoside phosphorylase
Purine metabolism in the parasite Plasmodium has been identified as a promising target for antimalarial therapies due to slight alterations around the active site when compared to mammalian purine nucleoside phosphorylase (PNP). PNP is part of a salvage pathway for the biosynthesis of purines, which are essential to parasite survival due to location of the organism, erthrocytes and Plasmodium itself not being able to produce purines de novo .
Reference Protein and Structure
- Sequence
-
Q8I3X4
 (2.4.2.1)
(2.4.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Plasmodium falciparum 3D7 (malaria parasite P. falciparum)

- PDB
-
2bsx
- Crystal structure of the Plasmodium falciparum purine nucleoside phosphorylase complexed with inosine
(2.0 Å)



- Catalytic CATH Domains
-
3.40.50.1580
 (see all for 2bsx)
(see all for 2bsx)
Enzyme Reaction (EC:2.4.2.1)
Enzyme Mechanism
Introduction
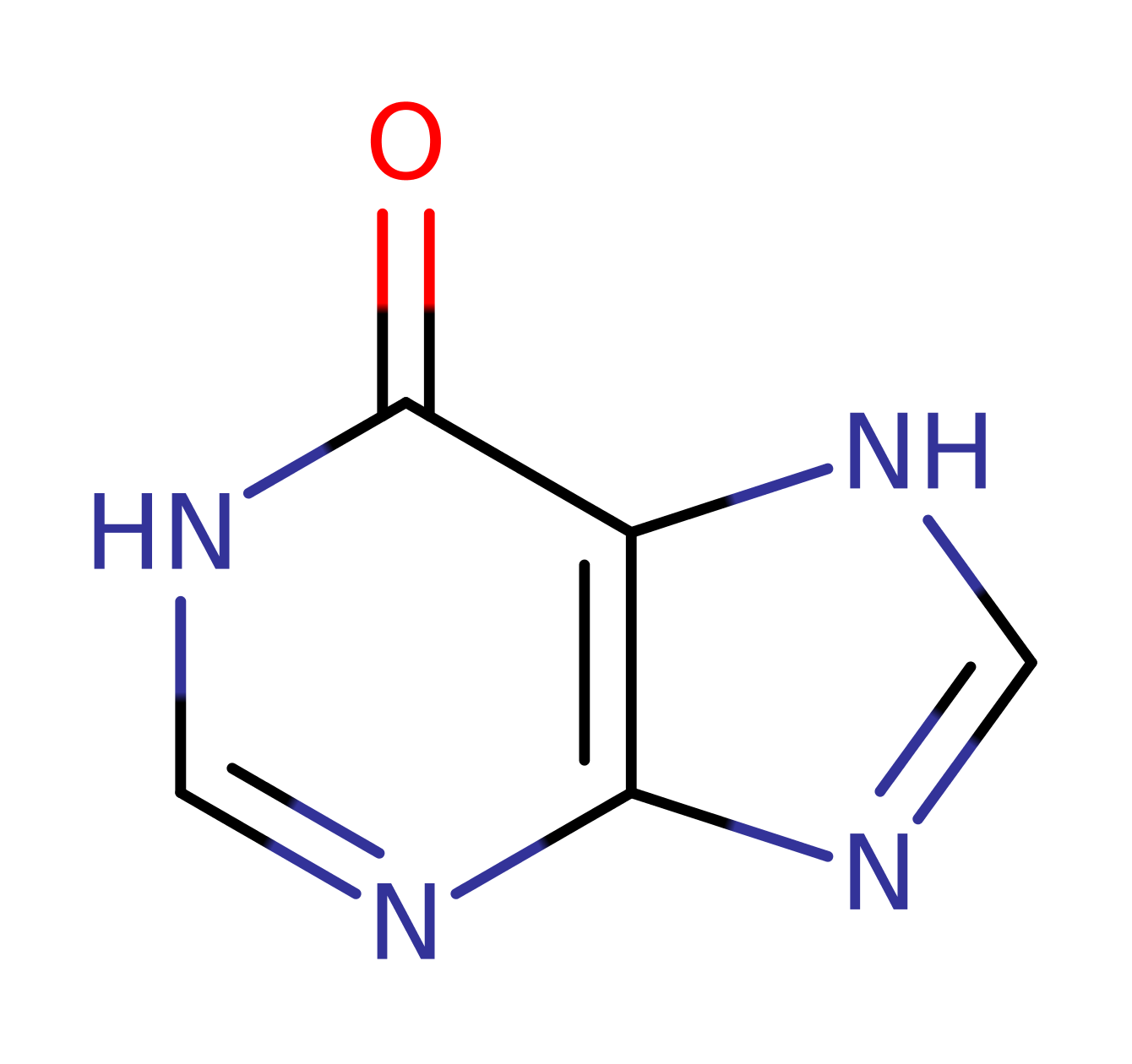
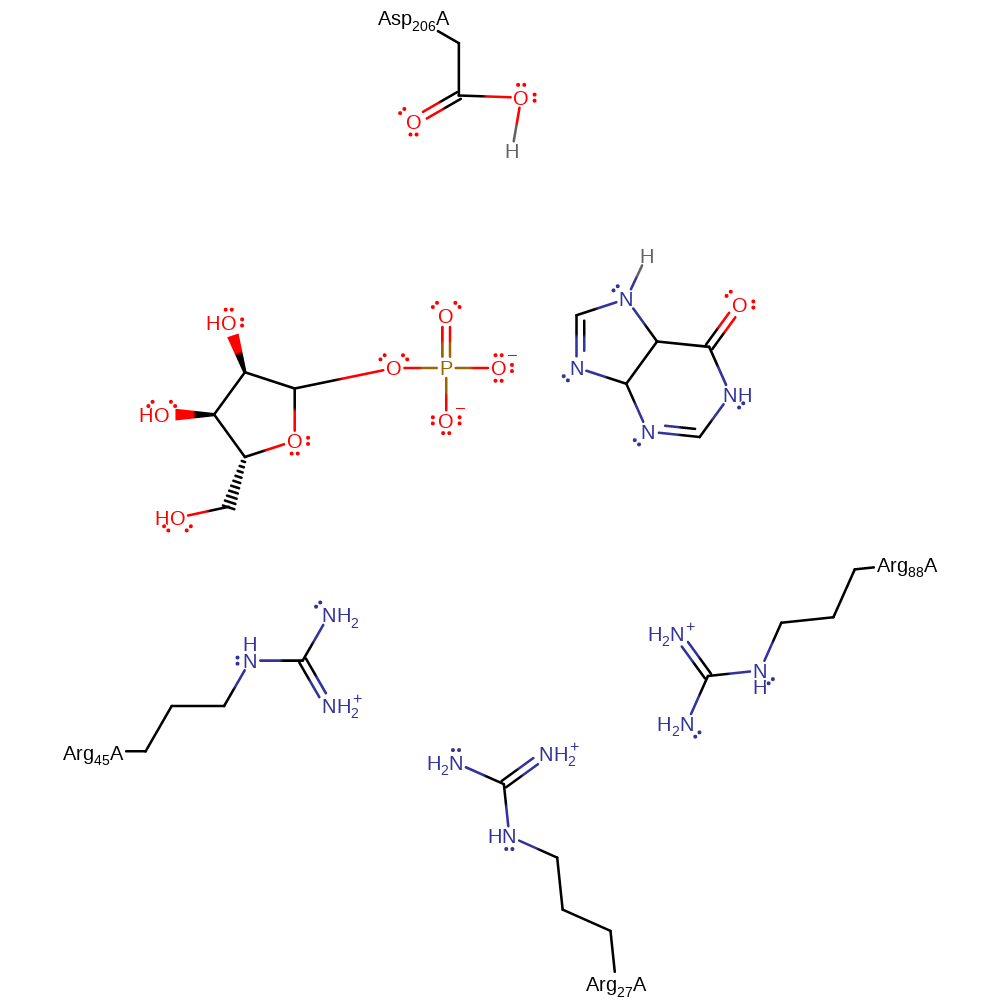
PNP is proposed to partake in a Sn1 nucleophilic substitution upon activation from conformational changes in the active site loop. It's substrate is inosine that has formed from prior deamination of adenosine by adenosine deaminase. Asp206 protonates N7 on the inosine group which facilitates glycosidic bond cleavage while Arg27 (alongside Arg45 and Arg88) stabilise the incoming oxygen nucleophile on a phosphate group. The reaction sees the formation of an oxocarbenium ion transition state which collapses to form ribose-phosphate and hypoxanthine which is a precursor to purine biosynthesis required by the parasite.
Catalytic Residues Roles
| UniProt | PDB* (2bsx) | ||
| Arg27 | Arg27A | Stabilises thus activates the phosphate anion for nucleophilic attack. | electrostatic stabiliser |
| Asp206 | Asp206A | Provides a proton to N7 of the purine as the glycosidic bond cleaves. | proton acceptor, proton donor |
| Arg88, Arg45 | Arg88A, Arg45A(AD) | Bind to and stabilise the phosphate ion via hydrogen bonds. | electrostatic stabiliser |
Chemical Components
heterolysis, charge delocalisation, proton transfer, elimination (not covered by the Ingold mechanisms), intermediate formation, overall reactant used, bimolecular nucleophilic addition, intermediate terminated, inferred reaction step, native state of enzyme regenerated, overall product formedReferences
- Chaikuad A et al. (2009), BMC Struct Biol, 9, 42-. Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases. DOI:10.1186/1472-6807-9-42. PMID:19575810.
- Madrid DC et al. (2008), J Biol Chem, 283, 35899-35907. Plasmodium falciparum purine nucleoside phosphorylase is critical for viability of malaria parasites. DOI:10.1074/jbc.M807218200. PMID:18957439.
- Schnick C et al. (2005), Acta Crystallogr D Biol Crystallogr, 61, 1245-1254. Structures ofPlasmodium falciparumpurine nucleoside phosphorylase complexed with sulfate and its natural substrate inosine. DOI:10.1107/s0907444905020251. PMID:16131758.
- Tahirov TH et al. (2004), J Mol Biol, 337, 1149-1160. Crystal Structure of Purine Nucleoside Phosphorylase from Thermus thermophilus. DOI:10.1016/j.jmb.2004.02.016. PMID:15046984.
- Bennett EM et al. (2003), J Biol Chem, 278, 47110-47118. Structural Basis for Substrate Specificity of Escherichia coli Purine Nucleoside Phosphorylase. DOI:10.1074/jbc.m304622200. PMID:12937174.
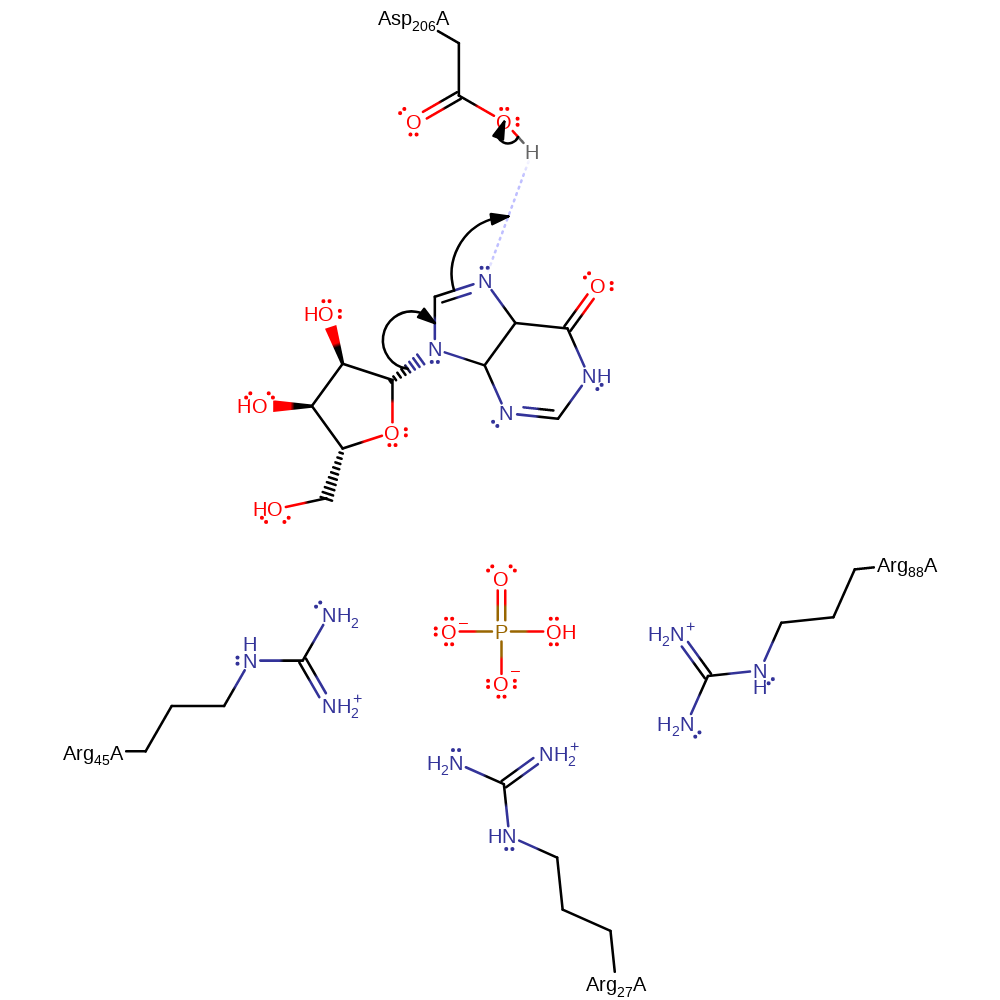
Step 1. Asp206 protonates the inosine base, promoting electron withdrawal to cleave the glycosidic bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg27A | electrostatic stabiliser |
| Arg45A(AD) | electrostatic stabiliser |
| Arg88A | electrostatic stabiliser |
| Asp206A | proton donor |
Chemical Components
heterolysis, charge delocalisation, proton transfer, elimination (not covered by the Ingold mechanisms), intermediate formation, overall reactant used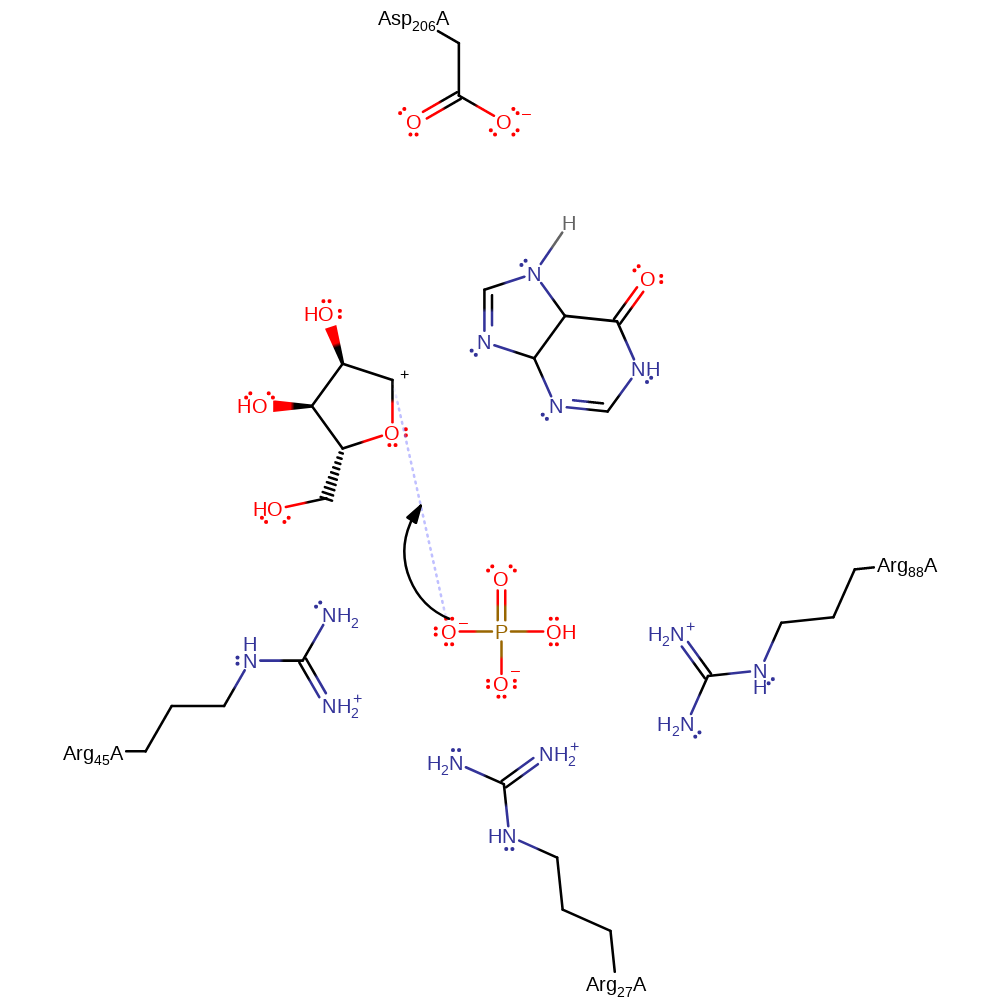
Step 2. Arg27 helps activate the phosphate group for nucleophilic attack on a carbon on the ribose ring by stabilising the negatively charged group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg27A | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp206A | proton acceptor |
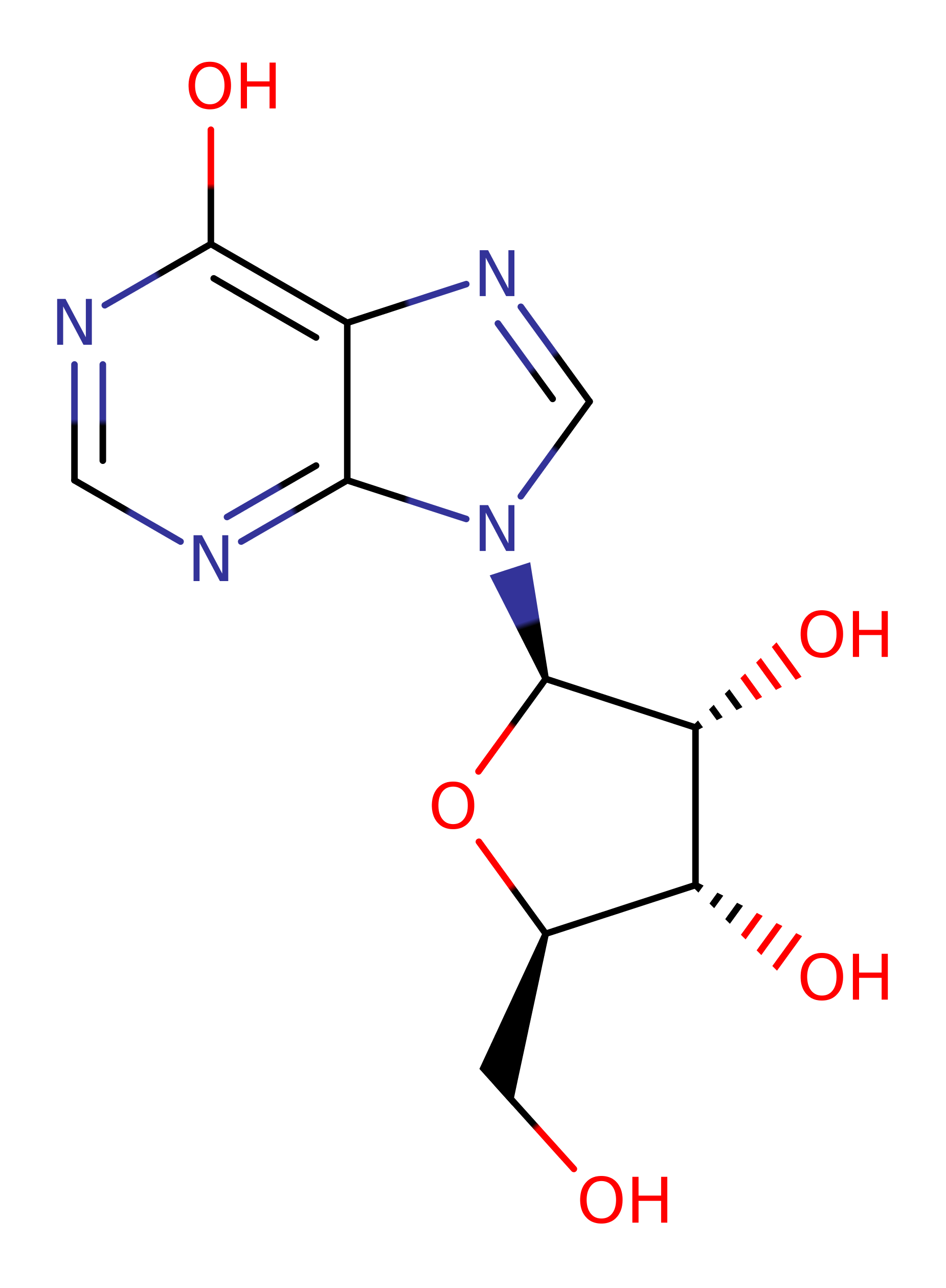

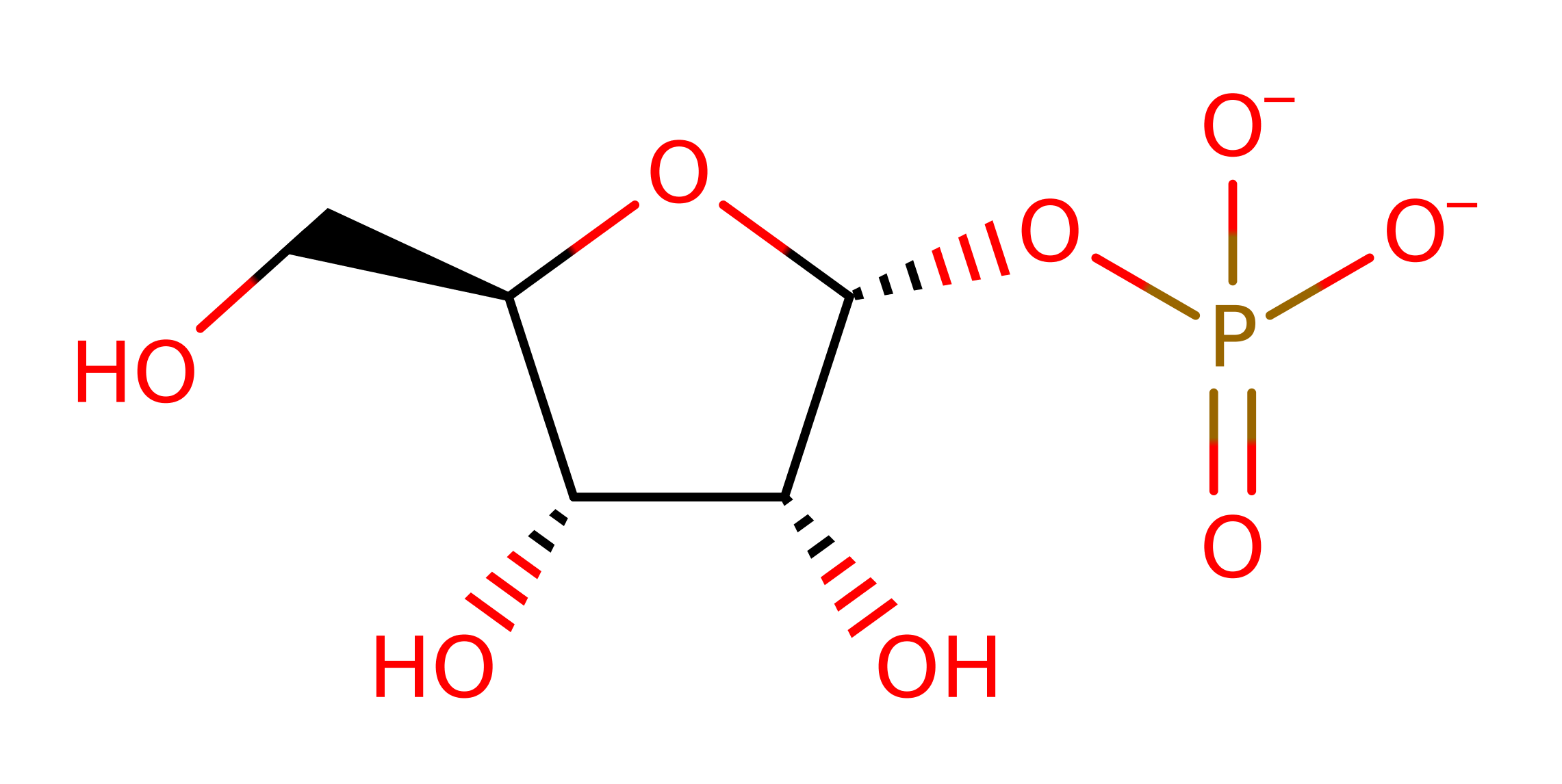

 Download:
Download: