Isocitrate dehydrogenase (NADP+)
Isocitrate dehydrogenase (ICD) is a crucial enzyme in the TCA cycle. Reversible inactivation of the enzyme by phosphorylation at Ser113 regulates metabolic switching from the TCA cycle to the glyoxylate pathway, which is adopted by bacteria, fungi and plants in the absence of a "complex" carbon source such as glucose. In humans, specific mutations within ICD are found in several forms of brain tumour, with some specificity between pathologies. Mutations in ICD are detected in almost all forms of secondary glioblastoma which form from lower-grade glioma, but they are rarely found in primary high-grade glioblastoma multiforme. A correlation between the presence of a ICD mutation and increased life expectancy from secondary glioblastoma has been suggested [PMID:19636000].
Reference Protein and Structure
- Sequence
-
P08200
 (1.1.1.42)
(1.1.1.42)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
5icd
- REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE
(2.5 Å)



- Catalytic CATH Domains
-
3.40.718.10
 (see all for 5icd)
(see all for 5icd)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:1.1.1.42)
Enzyme Mechanism
Introduction
Asp283AA activates is hydrogen bonded to both Lys230 and Tyr160. In this mechanism Asp activates the Lys to act as the general base in the first step and the Tyr to be the general acid in the last step. The hydride transfer from substrate to NADP+ is concerted with the initial deprotonation.
The second half of the reaction involves decarboxylation from C2, forming an enolate which is stabilised by the divalent cation and proximal positively charged residues. This intermediate then undergoes a rearrangement to form the final product with Tyr as the general acid.
Catalytic Residues Roles
| UniProt | PDB* (5icd) | ||
| Tyr160 | Tyr160A | Acts as a general acid/base and donates its proton to the final product. It is returned to its correct protonation state by abstracting a proton from Asp283. | hydrogen bond donor, proton acceptor, proton donor, proton relay, electrostatic stabiliser |
| Asp307 | Asp307A | Binds Mg(II) ion. | metal ligand |
| Asp283 | Asp283A(AA) | Acts as the general acid/base, performs the initial deprotonation of Lys230. Reprotonates Tyr160. | proton acceptor, electrostatic stabiliser, proton donor |
| Lys230 | Lys230A(AA) | Acts as a general acid/base, it is activated for its initial proton transfer by Asp230. It then abstracts a proton from the substrate. | proton acceptor, proton relay, electrostatic stabiliser, proton donor |
Chemical Components
proton transfer, hydride transfer, bimolecular elimination, aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed, intermediate formation, unimolecular elimination by the conjugate base, decarboxylation, intermediate collapse, assisted keto-enol tautomerisation, intermediate terminatedReferences
- Neves RPP et al. (2016), ACS Catal, 6, 357-368. Unveiling the Catalytic Mechanism of NADP+-Dependent Isocitrate Dehydrogenase with QM/MM Calculations. DOI:10.1021/acscatal.5b01928.
- Dang L et al. (2009), Nature, 462, 739-744. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. DOI:10.1038/nature08617. PMID:19935646.
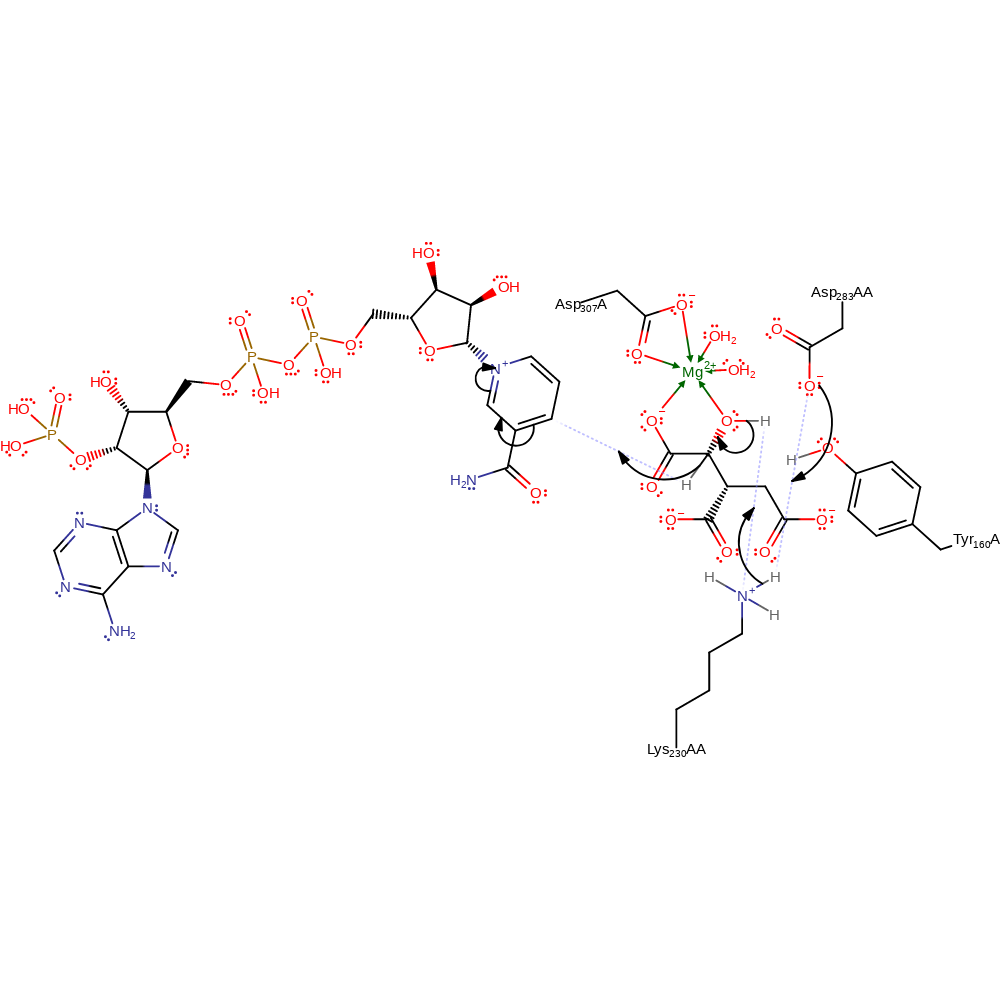
Step 1. Asp283AA deprotonates Lys230, which deprotonates the C2-OH, which causes the elimination of a hydride ion, which adds to NADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys230A(AA) | electrostatic stabiliser |
| Asp307A | metal ligand |
| Tyr160A | hydrogen bond donor, electrostatic stabiliser |
| Asp283A(AA) | proton acceptor |
| Lys230A(AA) | proton donor, proton acceptor, proton relay |
Chemical Components
proton transfer, hydride transfer, ingold: bimolecular elimination, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed, intermediate formation
Step 2. The intermediate decarboxylates, with concomitant double bond rearrangement.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor |
| Asp307A | metal ligand |
| Tyr160A | electrostatic stabiliser |
| Asp283A(AA) | electrostatic stabiliser |
| Lys230A(AA) | electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, overall product formed, decarboxylation, intermediate collapse, intermediate formation
Step 3. The oxyanion formed collapses, with concomitant deprotonation of Tyr160A, in turn re-protonated by Asp238AA
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor |
| Asp307A | metal ligand |
| Lys230A(AA) | electrostatic stabiliser |
| Tyr160A | proton donor, proton acceptor |
| Asp283A(AA) | proton donor |
| Tyr160A | proton relay |
Chemical Components
assisted keto-enol tautomerisation, intermediate terminated, overall product formed, proton transferIntroduction
Asp283AA acts as the base towards isocitrate C1-OH, initiating simultaneous hydride transfer to NADP+ while forming a carbonyl at this position. The second half of the reaction involves decarboxylation from C2, forming an enolate which is stabilised by the divalent cation and proximal positively charged residues. This intermediate then undergoes a rearrangement to form the C2-carbonyl and form alpha-ketoglutarate.
Catalytic Residues Roles
| UniProt | PDB* (5icd) | ||
| Tyr160 | Tyr160A | Activates Asp283 | hydrogen bond donor, electrostatic stabiliser |
| Asp307 | Asp307A | Binds Mg(II) ion. | metal ligand |
| Asp283 | Asp283A(AA) | Acts as the general acid/base, performs the initial deprotonation of the substrate. It is subsequently deprotonated by Lys230 in an inferred return step. | proton acceptor, electrostatic stabiliser, proton donor |
| Lys230 | Lys230A(AA) | Acts as a general acid/base, donating a proton to the final product. It is reprotonated by Asp283. | proton acceptor, electrostatic stabiliser, proton donor |
Chemical Components
bimolecular elimination, aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed, intermediate formation, hydride transfer, proton transfer, unimolecular elimination by the conjugate base, decarboxylation, intermediate collapse, assisted keto-enol tautomerisation, intermediate terminated, native state of enzyme regenerated, inferred reaction stepReferences
- Hurley JH et al. (1991), Biochemistry, 30, 8671-8678. Catalytic mechanism of NADP+-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes. DOI:10.1021/bi00099a026. PMID:1888729.
- Sanson M et al. (2009), J Clin Oncol, 27, 4150-4154. Isocitrate Dehydrogenase 1 Codon 132 Mutation Is an Important Prognostic Biomarker in Gliomas. DOI:10.1200/jco.2009.21.9832. PMID:19636000.
- Peng Y et al. (2008), Protein Sci, 17, 1542-1554. Structural studies ofSaccharomyces cerevesiaemitochondrial NADP-dependent isocitrate dehydrogenase in different enzymatic states reveal substantial conformational changes during the catalytic reaction. DOI:10.1110/ps.035675.108. PMID:18552125.
- Kim TK et al. (2005), Protein Sci, 14, 140-147. Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate dehydrogenase. DOI:10.1110/ps.041091805. PMID:15576556.
- Doyle SA et al. (2001), Biochemistry, 40, 4234-4241. Structural Basis for a Change in Substrate Specificity: Crystal Structure of S113E Isocitrate Dehydrogenase in a Complex with Isopropylmalate, Mg2+, and NADP†,‡. DOI:10.1021/bi002533q. PMID:11284679.
- Imada K et al. (1998), Structure, 6, 971-982. Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 å resolution: the role of Glu88 in the unique substrate-recognition mechanism. DOI:10.1016/s0969-2126(98)00099-9. PMID:9739088.
- Lee ME et al. (1995), Biochemistry, 34, 378-384. Mutational analysis of the catalytic residues lysine 230 and tyrosine 160 in the NADP+-dependent isocitrate dehydrogenase from Escherichia coli. DOI:10.1021/bi00001a046. PMID:7819221.
- Dean AM et al. (1993), Biochemistry, 32, 9302-9309. Kinetic mechanism of Escherichia coli isocitrate dehydrogenase. DOI:10.1021/bi00087a007. PMID:8369299.
- Thorsness PE et al. (1987), J Biol Chem, 262, 10422-10425. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. PMID:3112144.

Step 1. Asp283AA deprotonates the C2-OH, which causes the elimination of a hydride ion, which adds to NADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor, electrostatic stabiliser |
| Asp307A | metal ligand |
| Lys230A(AA) | electrostatic stabiliser |
| Asp283A(AA) | proton acceptor |
Chemical Components
ingold: bimolecular elimination, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed, intermediate formation, hydride transfer, proton transfer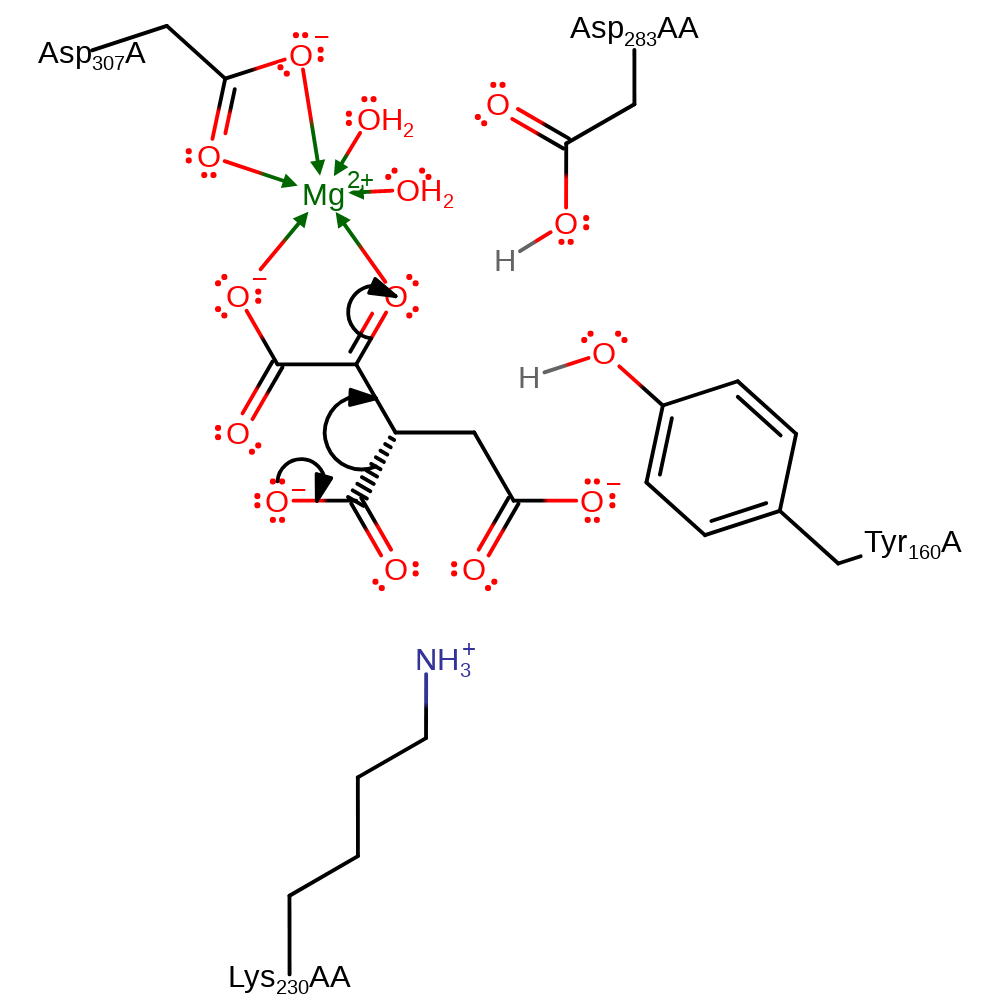
Step 2. The intermediate decarboxylates, with concomitant double bond rearrangement.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor |
| Tyr160A | electrostatic stabiliser |
| Asp307A | metal ligand |
| Asp283A(AA) | electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, overall product formed, decarboxylation, intermediate collapse, intermediate formation
Step 3. The oxyanion formed collapses, with concomitant deprotonation of Lys230B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor, electrostatic stabiliser |
| Asp283A(AA) | electrostatic stabiliser |
| Lys230A(AA) | proton donor |
Chemical Components
assisted keto-enol tautomerisation, proton transfer, overall product formed, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor |
| Asp307A | metal ligand |
| Tyr160A | electrostatic stabiliser |
| Lys230A(AA) | proton acceptor |
| Asp283A(AA) | proton donor |
Chemical Components
proton transfer, native state of enzyme regenerated, inferred reaction stepIntroduction
Asp283AA acts as the base towards isocitrate C1-OH, the charged intermediate is well stabilised by the Mg(II) ion, the hydride transfer from the intermediate to NADP+, forming a carbonyl at the C1 position, occurs in a separate step. The second half of the reaction involves decarboxylation from C2, forming an enolate which is stabilised by the divalent cation and proximal positively charged residues. This intermediate then undergoes a rearrangement to form the C2-carbonyl and form alpha-ketoglutarate.
Catalytic Residues Roles
| UniProt | PDB* (5icd) | ||
| Tyr160 | Tyr160A | Helps activate Asp283. | hydrogen bond donor, electrostatic stabiliser |
| Asp307 | Asp307A | Binds the Mg(II) ion. | metal ligand |
| Asp283 | Asp283A(AA) | Acts as a general acid/base, performing the initial deprotonation of the substrate. Lys230 deprotonates Asp283 to regenerate the starting state of the active site. | proton acceptor, electrostatic stabiliser, proton donor |
| Lys230 | Lys230A(AA) | Acts as a general acid/base, reprotonating the product. Lys230 deprotonates Asp283 ro regenerate the starting state of the active site. | proton acceptor, electrostatic stabiliser, proton donor |
Chemical Components
overall reactant used, intermediate formation, proton transfer, hydride transfer, bimolecular elimination, aromatic bimolecular nucleophilic addition, overall product formed, unimolecular elimination by the conjugate base, decarboxylation, intermediate collapse, assisted keto-enol tautomerisation, intermediate terminated, native state of enzyme regenerated, inferred reaction stepReferences
- Hurley JH et al. (1991), Biochemistry, 30, 8671-8678. Catalytic mechanism of NADP+-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes. DOI:10.1021/bi00099a026. PMID:1888729.

Step 1. Asp283AA deprotonates the C2-OH, the resulting anionic intermediate is well stabilised by the Mg(II) ion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys230A(AA) | electrostatic stabiliser |
| Asp307A | metal ligand |
| Tyr160A | hydrogen bond donor, electrostatic stabiliser |
| Asp283A(AA) | proton acceptor |
Chemical Components
overall reactant used, intermediate formation, proton transfer
Step 2. A hydride ion is eliminated from the intermediate and adds to NADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys230A(AA) | electrostatic stabiliser |
| Asp307A | metal ligand |
| Tyr160A | hydrogen bond donor, electrostatic stabiliser |
| Asp283A(AA) | electrostatic stabiliser |
Chemical Components
hydride transfer, ingold: bimolecular elimination, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, overall product formed, intermediate formation
Step 3. The intermediate decarboxylates, with concomitant double bond rearrangement.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor |
| Asp307A | metal ligand |
| Asp283A(AA) | electrostatic stabiliser |
| Tyr160A | electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, overall product formed, decarboxylation, intermediate collapse, intermediate formation
Step 4. The oxyanion formed collapses, with concomitant deprotonation of Lys230B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor, electrostatic stabiliser |
| Asp283A(AA) | electrostatic stabiliser |
| Lys230A(AA) | proton donor |
Chemical Components
assisted keto-enol tautomerisation, proton transfer, overall product formed, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr160A | hydrogen bond donor |
| Asp307A | metal ligand |
| Tyr160A | electrostatic stabiliser |
| Lys230A(AA) | proton acceptor |
| Asp283A(AA) | proton donor |





 Download:
Download: 
 Download:
Download: 
 Download:
Download: