Phenylalanine ammonia-lyase
Phenylalanine ammonia-lyase (PAL) is a plant enzyme which catalyses the non-oxidative elimination of ammonia from L-Phe to give trans-cinnamate. Trans-cinnamate is the precursor of numerous phenylpropanoid compounds and plays an important role in plant development and plant stress response. PAL may become a useful palliative for phenylketonuria as it has been shown to be able to convert excess Phe (toxic) in the blood into harmless compounds. It follows the same catalytic mechanism as tyrosine ammonia-lyase (TAL) for the deamination of L-tyrosine to (E)-4-coumarate.
Reference Protein and Structure
- Sequence
-
P24481
 (4.3.1.24)
(4.3.1.24)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Petroselinum crispum (parsley)

- PDB
-
1w27
- Phenylalanine ammonia-lyase (PAL) from Petroselinum crispum
(1.7 Å)



- Catalytic CATH Domains
-
1.10.275.10
 1.20.200.10
1.20.200.10  (see all for 1w27)
(see all for 1w27)
- Cofactors
- 2-[(1s)-1-aminoethyl]-1-carboxymethyl-5-hydroxy-4-methylimidazole (1)
Enzyme Reaction (EC:4.3.1.24)
Enzyme Mechanism
Introduction
In this mechanism, the reaction proceeds via an N-MIO intermediate rather than by an FC reaction. Ammonia is eliminated from the intermediate, forming the product and regenerating the cofactor.
Catalytic Residues Roles
| UniProt | PDB* (1w27) | ||
| Tyr110 | Tyr110A | Increases acidity of the of the intermediate to promote elimination | increase acidity |
| Tyr351 | Asp351(349)A | Act as a proton relay to cause the elimination of ammonia. | proton relay, proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic addition, cofactor used, overall reactant used, intermediate formation, intramolecular elimination, overall product formed, native state of cofactor regenerated, intermediate terminated, proton relayReferences
- Weiser D et al. (2015), Chembiochem, 16, 2283-2288. Phenylalanine Ammonia-Lyase-Catalyzed Deamination of an Acyclic Amino Acid: Enzyme Mechanistic Studies Aided by a Novel Microreactor Filled with Magnetic Nanoparticles. DOI:10.1002/cbic.201500444. PMID:26345352.
- Pinto GP et al. (2015), Arch Biochem Biophys, 582, 107-115. New insights in the catalytic mechanism of tyrosine ammonia-lyase given by QM/MM and QM cluster models. DOI:10.1016/j.abb.2015.03.002. PMID:25772386.

Step 1. The amine group of the substrate performs a nucleophilic attack on the C=C bond of the cofactor, forming a substrate cofactor intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, cofactor used, overall reactant used, intermediate formation
Step 2. Ty351 acts as a proton relay this leads to the elimination of ammonia, the formation of the product and the regeneration of the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp351(349)A | proton relay |
| Tyr110A | increase acidity |
| Asp351(349)A | proton acceptor, proton donor |
Chemical Components
ingold: intramolecular elimination, overall product formed, native state of cofactor regenerated, intermediate terminated, proton relayIntroduction
This mechanism proposal involves only a single step. There is no covalent bond between the cofactor and the substrate and the cofactor only acts to stabilize the transition state. The elimination of ammonia is facilitated by acid/base catalysis of the two tyrosine residues. This catalytic path is supported by QM/MM calculations.
Catalytic Residues Roles
| UniProt | PDB* (1w27) | ||
| Tyr110 | Tyr110A | Acts as an acid to make the ammonia a better leaving group | promote heterolysis, proton donor |
| Tyr351 | Asp351(349)A | Deprotonates the substrate to facilitate the elimination of ammonia | proton acceptor |
Chemical Components
overall reactant used, overall product formed, bimolecular elimination, proton transferReferences
- Weiser D et al. (2015), Chembiochem, 16, 2283-2288. Phenylalanine Ammonia-Lyase-Catalyzed Deamination of an Acyclic Amino Acid: Enzyme Mechanistic Studies Aided by a Novel Microreactor Filled with Magnetic Nanoparticles. DOI:10.1002/cbic.201500444. PMID:26345352.
- Pinto GP et al. (2015), Arch Biochem Biophys, 582, 107-115. New insights in the catalytic mechanism of tyrosine ammonia-lyase given by QM/MM and QM cluster models. DOI:10.1016/j.abb.2015.03.002. PMID:25772386.

Step 1. Tyr351 deprotonates the substrate and Tyr110 protonates the amino group, this makes it a better leaving group allowing the E2 reaction to occur.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr110A | promote heterolysis |
| Tyr110A | proton donor |
| Asp351(349)A | proton acceptor |
Chemical Components
overall reactant used, overall product formed, ingold: bimolecular elimination, proton transferIntroduction
The electrophile in the mechanism is thought to be formed by the autocatalytic formation of 3,5-dihydro-5-methyldiene-4H-imidazole-4-one group (MIO) from the tripeptide Ala202-Ser203-Gly204. The MIO group is formed by cyclisation as a result of two water elimination steps. The sp3 conformation at Gly204 amide N increases the electrophicity of the Ser203 beta-C atom. In the reaction mechanism, an electron pair of the substrate phenyl ring attacks the beta-C of Ser203 of the MIO causing the MIO to become aromatic. The conversion of MIO to the aromatic state changes the conformation of Gly204 amide-N from sp3 to sp2, causing a small peptide displacement. The Ser203 oxygen anion produced stabilises the positively charged sigma-complex of the substrate phenyl group. The positive charge is also stabilised by an interaction with the pi-electrons of Phe400. The electron-deficient phenyl ring of L-Phe renders the two hydrogen atoms at the beta-C atom of the substrate acidic. Tyr351' from another subunit abstracts the pro-S beta-hydrogen from the substrate. The rearrangement of the Phe-MIO adduct eliminates the amino group and regenerates MIO. The resulting ammonia is released into the solvent after the trans-cinnamate has diffused away. The proton abstracted by Tyr351' can be easily released to the bulk solvent to regenerate the enzyme.
Catalytic Residues Roles
| UniProt | PDB* (1w27) | ||
| Tyr351 | Asp351(349)A | Tyr351' acts as a base, abstracting a beta-H from the substrate.The proton is released into the bulk solvent. | proton acceptor, proton donor |
| Phe400 | Gly400(398)A | Phe400 stabilises the positively charged sigma-complex intermediate to prevent the removal of the proton in the ortho-position of the aromatic ring. | electrostatic stabiliser |
Chemical Components
aromatic bimolecular electrophilic addition, intermediate formation, overall reactant used, cofactor used, proton transfer, intramolecular elimination, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, intermediate terminatedReferences
- Ritter H et al. (2004), Plant Cell, 16, 3426-3436. Structural Basis for the Entrance into the Phenylpropanoid Metabolism Catalyzed by Phenylalanine Ammonia-Lyase. DOI:10.1105/tpc.104.025288. PMID:15548745.
- Weiser D et al. (2015), Chembiochem, 16, 2283-2288. Phenylalanine Ammonia-Lyase-Catalyzed Deamination of an Acyclic Amino Acid: Enzyme Mechanistic Studies Aided by a Novel Microreactor Filled with Magnetic Nanoparticles. DOI:10.1002/cbic.201500444. PMID:26345352.
- Röther D et al. (2002), Eur J Biochem, 269, 3065-3075. An active site homology model of phenylalanine ammonia-lyase fromP. crispum. DOI:10.1046/j.1432-1033.2002.02984.x. PMID:12071972.

Step 1. The phenyl ring attacks the C=C bond of the MIO cofactor forming a substrate cofactor intermediate. The resulting carbocation is stabilized by Phe400.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly400(398)A | electrostatic stabiliser |
Chemical Components
ingold: aromatic bimolecular electrophilic addition, intermediate formation, overall reactant used, cofactor used
Step 2. Tyr351 deprotonates the beta carbon of the substrate, neutralizing the carbocation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly400(398)A | electrostatic stabiliser |
| Asp351(349)A | proton acceptor |
Chemical Components
proton transfer
Step 3. Ammonia is eliminated from the substrate and the cofactor is regenerated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp351(349)A | proton donor |

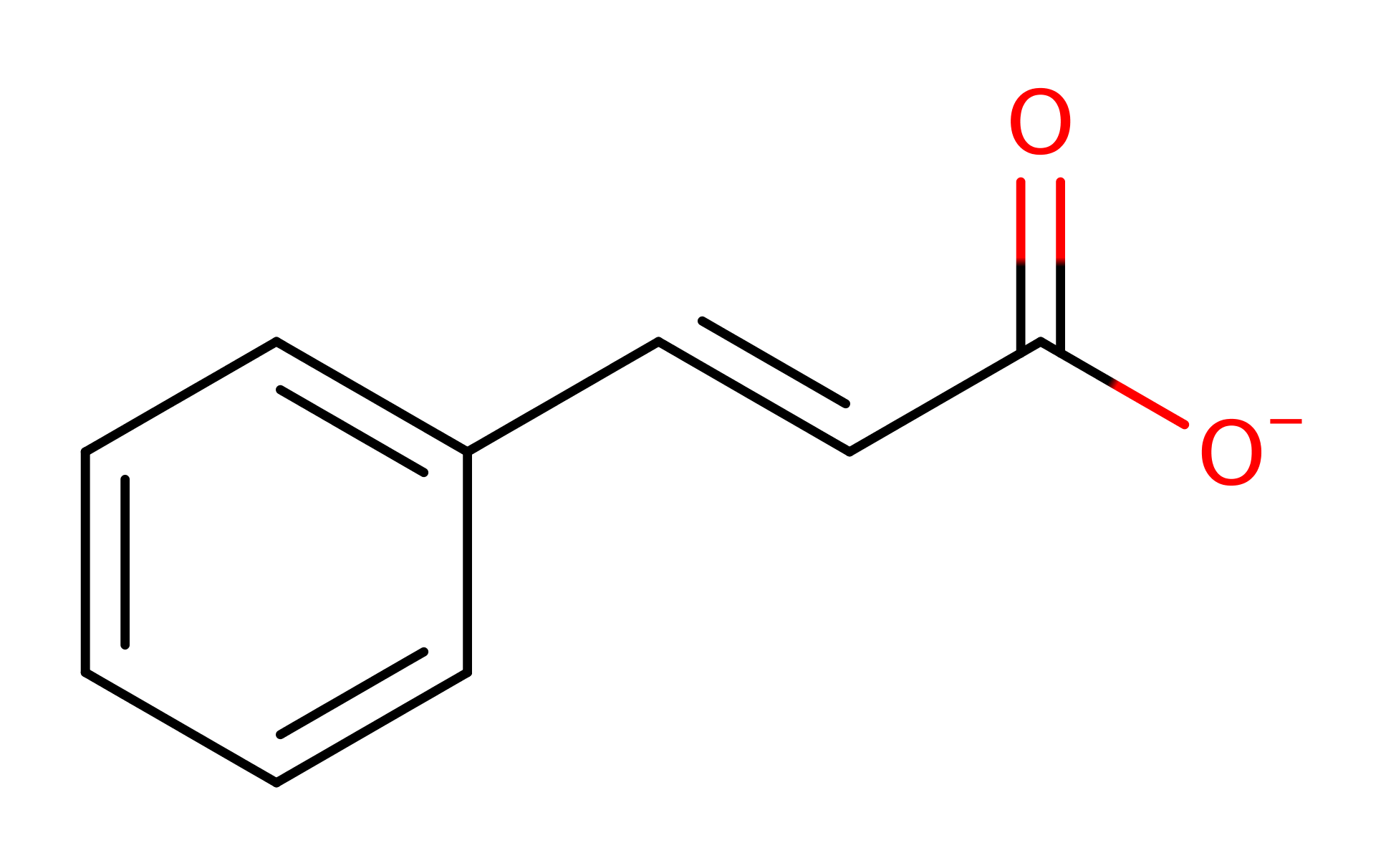

 Download:
Download:  Download:
Download:  Download:
Download: