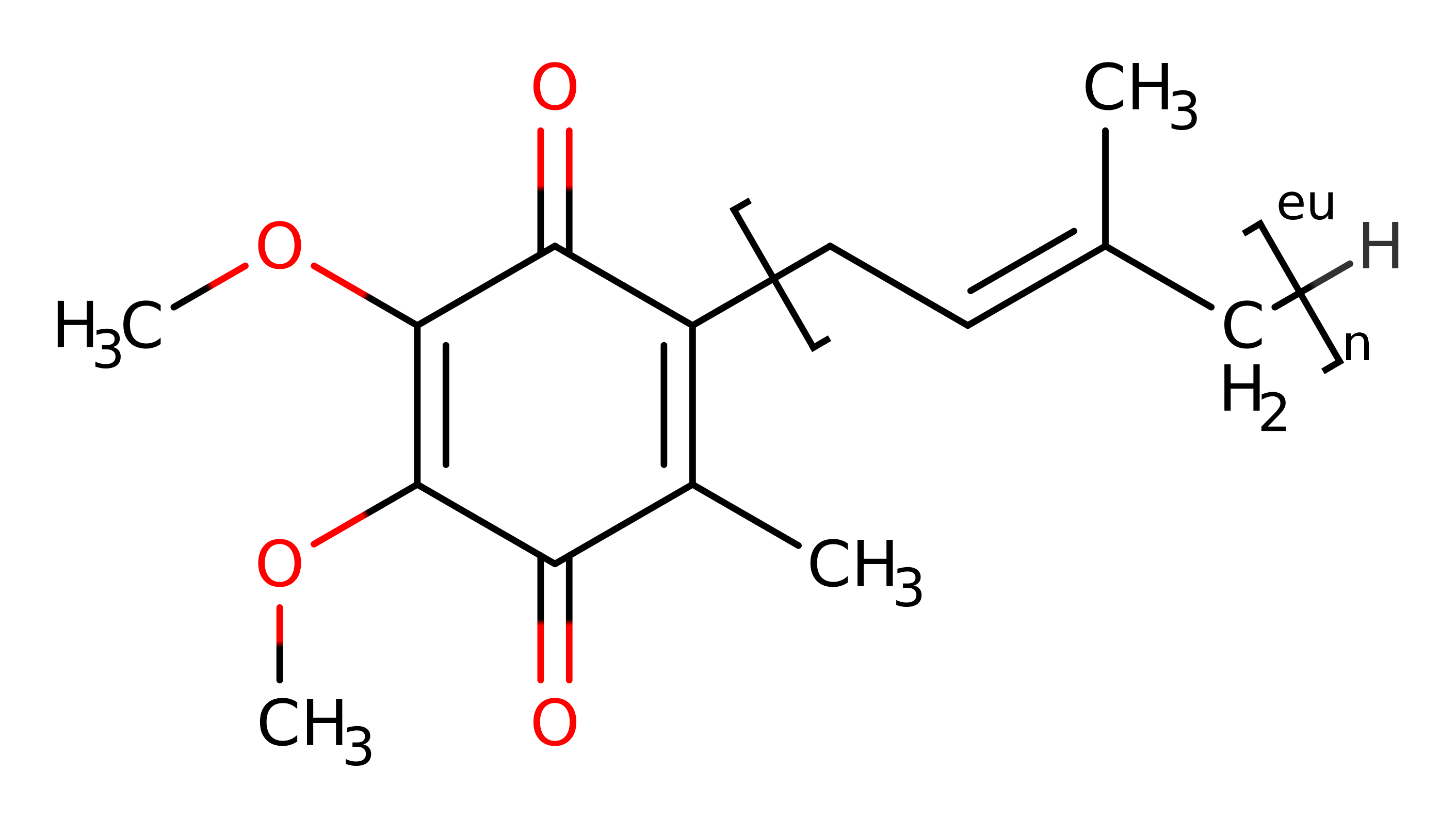
Ubiquinol oxidase (H+-transporting)
The cytochrome bo3 ubiquinol oxidase from E. coli is a terminal enzyme in the respiratory chain, oxidising the membrane-soluble ubiquinol-8 and reducing molecular dioxygen, and pumping four protons across the membrane to generate a proton gradient that is used to drive ATP synthesis. Ubiquinol oxidase is homologous to cytochrome c oxidase; both have four subunits, the same redox mechanism, the same electron transfer pathway and conserved proton pumping channels.
Reference Protein and Structure
- Sequences
-
P0ABI8
 (7.1.1.3)
(7.1.1.3)
P0ABJ1
P0ABJ3
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1fft
- The structure of ubiquinol oxidase from Escherichia coli
(3.5 Å)



- Catalytic CATH Domains
-
1.20.210.10
 (see all for 1fft)
(see all for 1fft)
- Cofactors
- Copper(2+) (1), Copper(2+) (1), Heme b (1), Ferroheme o (1)
Enzyme Reaction (EC:7.1.1.3)
Enzyme Mechanism
Introduction
The bo3 ubiquinol oxidase has a tightly bound ubiquinol at the QH (high affinity) site, low-spin protoheme b (heme b) and a redox active binuclear centre comprising a copper ion (CuB) and an O-type heme (heme o3). Tyr 288 is near CuB and heme o3, and has a low pKa due to a covalent bond with His 284. In the resting (oxidised or O) state, the heme o3 contains Fe(III) and CuB is Cu(II). A hydroxide ion is bound by Fe and CuB.
A fully reduced ubiquinol docks in a low affinity QL site. From here two electrons are transferred to the tightly bound ubiquinone at the QH site. An electron is transferred from the QH site (via Met 79, Phe 103, heme b, His 421, Phe 420 and His 419) to the binuclear centre, giving a semi-reduced E state. This occurs at the same time as a proton is accepted by the hydroxide to give a water, which leaves. The semiubiquinone at QH is a neutral radical, stabilised by hydrogen bonding between semiubiquinone atom O1 (which is protonated) and Asp 75 and Arg 71. The second electron is transferred from the semiquinone at QH to the binuclear centre (via the same route as the first electron) to give the fully reduced R state, in which the binuclear centre contains Fe(II) and Cu(I). Molecular oxygen then binds to Fe(II), giving a short-lived A state. The next intermediate depends on whether there is an electron available on heme b (which will be delivered by a second ubiquinol docking at site QL, via the same route as before):
(a) If there is an electron available on heme b, four electrons (one from heme b, one from CuB, two from heme o3) reduce oxygen to yield a oxo-ferryl adduct with Fe(IV)=O and Cu(II) (the PR state). Water is produced, with the protons from a pool of water above the active site and/or Tyr 288.
(b) If there is no electron available on heme b, four electrons (one from CuB, two from heme o3, one from Tyr 288) reduce oxygen to yield a oxo-ferryl adduct with Fe(IV)=O, Cu(II) and a Tyr 288 radical (the PM state). Water is produced, with the protons from a pool of water above the active site and Tyr 288.
Both the P states decay to the F state (by protonation of Tyr 288 for PR, and reduction and protonation for PM) which also has Fe(IV)=O and Cu(II) but a regenerated Tyr 288. The second electron from the second QL ubiquinol is delivered to give the resting O state with Fe(III), Cu(II), hydroxide and protonated Tyr 288. The protons for water production and the associated proton pumping are sourced from the cytosolic side of the membrane.
There are two channels, homologous to the K and D channels of the better studied cytochrome c oxidase:
- K channel - from Ser 315, via Ser 299, Lys 362, Thr 359 and the heme o3 hydroxyethylfarnesyl tail, to Tyr 288
- D channel - from Asp 135, via Asn 124, Thr 211, Asn 142, Asn 124, Tyr 61, Thr 204, Ser 145, Thr 201 and Thr 149, to Glu 286.
The K channel is used for two protons required to move from the O state to the R state; the D channel is used for the six other protons required for movement from the A state through to the O state. The mechanism for coupling oxygen reduction and proton pumping is not known and has long been a topic of debate.
Catalytic Residues Roles
| UniProt | PDB* (1fft) | ||
| Arg71 | Arg71A | Arg 71 stabilises the transient semiquinone at the QH site. | electrostatic stabiliser |
| Asp75 | Asp75A | Asp 75 stabilises the transient semiquinone at the QH site. | electrostatic stabiliser |
| His284 | His284A | His 284, a ligand to CuB, is covalently bound to Tyr 288. This brings Tyr 288 into a position with lower pKa. | modifies pKa, metal ligand |
| Thr201, Tyr61, Asn142, Asn124, Asp135, Ser145, Thr149, Thr204, Thr211, Glu286 | Thr201A, Tyr61A, Asn142A, Asn124A, Asp135A, Ser145A, Thr149A, Thr204A, Thr211A, Glu286A | Part of the D-channel involved in proton transfer. | proton shuttle (general acid/base) |
| Tyr288 | Tyr288A | Tyr 88 donates an electron and / or a proton to oxygen to reduce it to water. Tyr 288 is also involved in the K channel. |
proton shuttle (general acid/base), electron shuttle |
| Thr359 | Thr359A | Part of the K-channel involved in proton transfer to the binuclear centre. | proton shuttle (general acid/base) |
| Ser299, Ser315, Lys362 | Ser299A, Ser315A, Lys362A | Part of the K-channel involved in proton transfer. | proton shuttle (general acid/base) |
| Met79, Phe103, Phe420, His419, His421 | Met79A, Phe103A, Phe420A, His419A, His421A | Part of the electron transfer chain involved in passing electrons from the ubiquinols to the binuclear centre. | electron shuttle |
Chemical Components
References
- Abramson J et al. (2000), Nat Struct Biol, 7, 910-917. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. DOI:10.1038/82824. PMID:11017202.
- Szundi I et al. (2014), Biochemistry, 53, 5393-5404. Kinetics and intermediates of the reaction of fully reduced Escherichia coli bo₃ ubiquinol oxidase with O₂. DOI:10.1021/bi500567m. PMID:25076393.
- Egawa T et al. (2011), Biochim Biophys Acta, 1807, 1342-1348. Differential effects of glutamate-286 mutations in the aa(3)-type cytochrome c oxidase from Rhodobacter sphaeroides and the cytochrome bo(3) ubiquinol oxidase from Escherichia coli. DOI:10.1016/j.bbabio.2011.06.001. PMID:21684251.
- Yap LL et al. (2007), J Biol Chem, 282, 8777-8785. Characterization of mutants that change the hydrogen bonding of the semiquinone radical at the QH site of the cytochrome bo3 from Escherichia coli. DOI:10.1074/jbc.M611595200. PMID:17267395.
- Yap LL et al. (2006), J Biol Chem, 281, 16879-16887. Characterization of the Exchangeable Protons in the Immediate Vicinity of the Semiquinone Radical at the QH Site of the Cytochrome bo3 from Escherichia coli. DOI:10.1074/jbc.m602544200. PMID:16624801.
- Brunori M et al. (2005), J Inorg Biochem, 99, 324-336. Cytochrome oxidase, ligands and electrons. DOI:10.1016/j.jinorgbio.2004.10.011. PMID:15598510.
- Tomson F et al. (2002), Biochemistry, 41, 14383-14390. Direct Infrared Detection of the Covalently Ring Linked His−Tyr Structure in the Active Site of the Heme−Copper Oxidases†. DOI:10.1021/bi026370c. PMID:12450405.
- Konstantinov AA et al. (1997), Proc Natl Acad Sci U S A, 94, 9085-9090. The roles of the two proton input channels in cytochrome c oxidase from Rhodobacter sphaeroides probed by the effects of site-directed mutations on time-resolved electrogenic intraprotein proton transfer. DOI:10.1073/pnas.94.17.9085. PMID:9256439.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| His419A | electron shuttle |
| Lys362A | proton shuttle (general acid/base) |
| Asp135A | proton shuttle (general acid/base) |
| Thr211A | proton shuttle (general acid/base) |
| Thr359A | proton shuttle (general acid/base) |
| Ser315A | proton shuttle (general acid/base) |
| Ser299A | proton shuttle (general acid/base) |
| Asn142A | proton shuttle (general acid/base) |
| Tyr61A | proton shuttle (general acid/base) |
| Thr201A | proton shuttle (general acid/base) |
| Ser145A | proton shuttle (general acid/base) |
| Met79A | electron shuttle |
| Asn124A | proton shuttle (general acid/base) |
| Phe420A | electron shuttle |
| Phe103A | electron shuttle |
| Glu286A | proton shuttle (general acid/base) |
| Thr149A | proton shuttle (general acid/base) |
| His421A | electron shuttle |
| Thr204A | proton shuttle (general acid/base) |
| Asp75A | electrostatic stabiliser |
| Arg71A | electrostatic stabiliser |
| Tyr288A | electron shuttle, proton shuttle (general acid/base) |
| His284A | metal ligand, modifies pKa |