4a-hydroxytetrahydrobiopterin dehydratase
4a-Hydroxy-tetrahydrobiopterin dehydratase/DCoH is a bifunctional protein. In the cytoplasm it is an enzyme required for the regeneration of tetrahydrobiopterin, an essential cofactor for phenylalanine hydroxylase. In the nucleus it functions as a transcriptional coactivator by forming a 2:2 heterotetramer with the hepatic nuclear factor HNF1alpha (HNF1).
The enzymatic catalytic form is a tetramer whereas the mode of binding to the nuclear factor is as a heterotetramer. The enzyme catalyses the dehydration of 4a-carbinolamine biopterin cofactor, a substrate used by the aromatic acid hydroxylases and NO synthase. Mutations of DcoHin humans have been associated with elevated levels of phenylalanine and the excretion of large amounts of 7-substituted pterins.
Reference Protein and Structure
- Sequence
-
P61459
 (4.2.1.96)
(4.2.1.96)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rattus norvegicus (Norway rat)

- PDB
-
1dco
- DCOH, A BIFUNCTIONAL PROTEIN-BINDING TRANSCRIPTIONAL COACTIVATOR
(2.3 Å)



- Catalytic CATH Domains
-
3.30.1360.20
 (see all for 1dco)
(see all for 1dco)
Enzyme Reaction (EC:4.2.1.96)
Enzyme Mechanism
Introduction
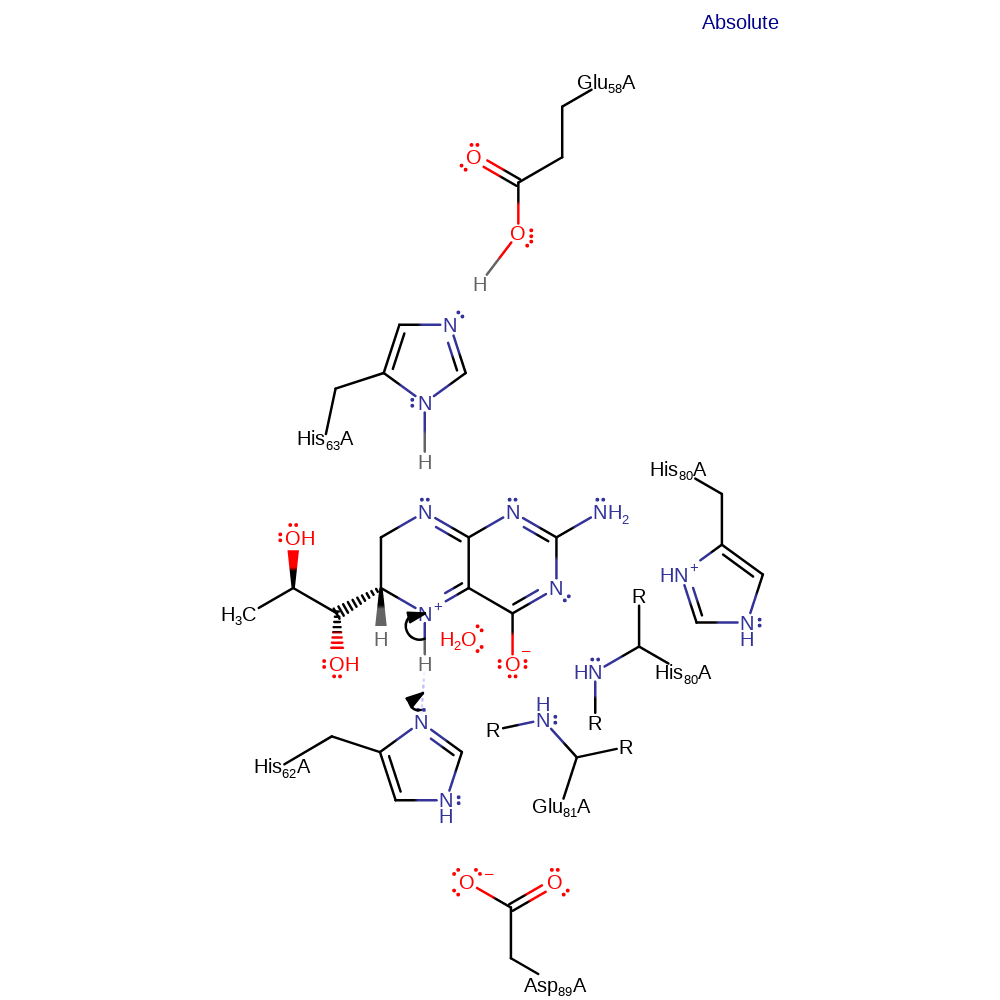
His63, activated by Glu58 abstracts the N(8) proton. The anion resides on the C-OH group in an enolate intermediate. The elimination of water from C4a is stereospecific, with His62 and His80 acting as general acids towards the 4a(R) hydroxyl and 4a(S) hydroxyl respectively. The residues then abstract a proton from N(5), relieving the atoms positive charge. This leads to the collapse of the anionic tetrahedral intermediate which has been stabilised by the presence of an oxyanion hole, resulting in the regeneration of the carbonyl at C(4) and reprotonation of N(8) by His63.
Catalytic Residues Roles
| UniProt | PDB* (1dco) | ||
| Glu81 (main-N) | Glu81A (main-N) | The backbone amide forms an oxyanion hole, stabilising the oxyanion intermediate. | hydrogen bond donor, electrostatic stabiliser |
| His80 | His80A | Acts as the general acid/base in the 4a(S),6(R) diastereoisomer. Putatively identified as a stabilising influence in the 4a(R),6(R) diastereoisomer reaction. | electrostatic stabiliser |
| Glu58 | Glu58A | Glu58 activates His63 through a hydrogen bond to act as a general base towards the N(5) atom of the substrate. | proton acceptor, hydrogen bond acceptor, electrostatic stabiliser, proton donor |
| His63 | His63A | The residue acts as a general base towards the N(5) atom of the substrate which leads to the formation of a tetrahedral oxyanion at C4. In the last step of the reaction, the residue reprotonates N(5), giving the quinoid dihydrobiopterin product. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Asp89 | Asp89A | Activates His62 to act as a general acid/base during the reaction. | hydrogen bond acceptor, electrostatic stabiliser |
| His80 (main-N) | His80A (main-N) | The imidazole side chain acts as a general acid/base towards the 4a(S) stereoisomer of the substrate. The backbone amide is implicated in stabilising the oxyanion intermediate. | hydrogen bond donor, electrostatic stabiliser |
| His62 | His62A | The imidazole side chain acts as a general acid/base towards the 4a(R) stereoisomer of the substrate. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
Chemical Components
proton transfer, overall reactant used, intermediate formation, unimolecular elimination by the conjugate base, overall product formed, intermediate collapse, dehydration, intermediate terminated, native state of enzyme regeneratedReferences
- Rebrin I et al. (1998), Biochemistry, 37, 11246-11254. Stereospecificity and Catalytic Function of Histidine Residues in 4a-Hydroxy-tetrahydropterin Dehydratase/DCoH†. DOI:10.1021/bi980663h. PMID:9698371.
- Hevel JM et al. (2008), Arch Biochem Biophys, 477, 356-362. Determinants of oligomerization of the bifunctional protein DCoHα and the effect on its enzymatic and transcriptional coactivator activities. DOI:10.1016/j.abb.2008.06.023. PMID:18644344.
- Cameron S et al. (2008), Mol Biochem Parasitol, 158, 131-138. Crystal structures of Toxoplasma gondii pterin-4a-carbinolamine dehydratase and comparisons with mammalian and parasite orthologues. DOI:10.1016/j.molbiopara.2007.12.002. PMID:18215430.
- Hevel JM et al. (2006), Mol Genet Metab, 88, 38-46. Can the DCoHα isozyme compensate in patients with 4a-hydroxy-tetrahydrobiopterin dehydratase/DCoH deficiency? DOI:10.1016/j.ymgme.2005.11.014. PMID:16423549.
- Köster S et al. (1998), Biol Chem, 379, 1427-1432. Pterin-4a-carbinolamine dehydratase from Pseudomonas aeruginosa: characterization, catalytic mechanism and comparison to the human enzyme. PMID:9894810.
- Cronk JD et al. (1996), Protein Sci, 5, 1963-1972. High-resolution structures of the bifunctional enzyme and transcriptional coactivator DCoH and its complex with a product analogue. DOI:10.1002/pro.5560051002. PMID:8897596.
- Koster S et al. (1996), Eur J Biochem, 241, 858-864. Location of the Active Site and Proposed Catalytic Mechanism of Pterin-4A-Carbinolamine Dehydratase. DOI:10.1111/j.1432-1033.1996.00858.x.
- Endrizzi J et al. (1995), Science, 268, 556-559. Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator. DOI:10.1126/science.7725101.
- Rebrin I et al. (1995), Biochemistry, 34, 5801-5810. Catalytic characterization of 4a-hydroxytetrahydropterin dehydratase. PMID:7727440.
- Citron BA et al. (1992), Proc Natl Acad Sci U S A, 89, 11891-11894. Identity of 4a-carbinolamine dehydratase, a component of the phenylalanine hydroxylation system, and DCoH, a transregulator of homeodomain proteins. PMID:1465414.

Step 1. His63 deprotonates the substrate initiating a double bond rearrangement that forms an oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu58A | hydrogen bond acceptor |
| His62A | hydrogen bond donor |
| His63A | hydrogen bond acceptor, hydrogen bond donor |
| His80A (main-N) | hydrogen bond donor |
| Glu81A (main-N) | hydrogen bond donor |
| Asp89A | hydrogen bond acceptor, electrostatic stabiliser |
| His80A | electrostatic stabiliser |
| His63A | proton donor, proton acceptor |
| Glu58A | proton acceptor |
| His63A | proton relay |
Chemical Components
proton transfer, overall reactant used, intermediate formation
Step 2. The substrate eliminates water, which obtains its proton from His62.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu58A | hydrogen bond acceptor, electrostatic stabiliser |
| His62A | hydrogen bond donor |
| His63A | hydrogen bond donor |
| His80A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Glu81A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asp89A | hydrogen bond acceptor, electrostatic stabiliser |
| His80A | electrostatic stabiliser |
| His62A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, overall product formed, intermediate collapse, intermediate formation, dehydrationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu58A | hydrogen bond acceptor, electrostatic stabiliser |
| His62A | hydrogen bond acceptor, hydrogen bond donor |
| His63A | hydrogen bond donor |
| His80A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Glu81A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asp89A | hydrogen bond acceptor |
| His80A | electrostatic stabiliser |
| His62A | proton acceptor |
Chemical Components
proton transfer, intermediate formation
Step 4. The oxyanion collapses, initiating double bond rearrangement that results in the deprotonation of His63 and the formation of the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu58A | hydrogen bond acceptor, electrostatic stabiliser |
| His62A | hydrogen bond donor |
| His63A | hydrogen bond donor |
| His80A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Glu81A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asp89A | hydrogen bond acceptor, electrostatic stabiliser |
| His63A | proton acceptor |
| Glu58A | proton donor |
| His63A | proton donor, proton relay |



 Download:
Download: