Quercetin 2,3-dioxygenase
Quercetin 2,3-dioxygenase is a copper-dependent enzyme that catalyses the reaction of dioxygen with quercin (3,5,7,3',4'-pentahydroxy flavone). Dioxygenases are enzymes that catalyses the incorporation of both atoms of molecular oxygen into organic substrates, most notably during the degradation of aromatic compounds. They typically use a metal ion to circumvent the spin barrier that prevents the direct reaction of the triplet ground state of dioxygen with singlet-state organic compounds. In most of the well-studied dioxygenases, the metal used is iron; quercetin 2,3-dioxygenase is the only known member of this family to use copper.
Reference Protein and Structure
- Sequence
-
Q7SIC2
 (1.13.11.24)
(1.13.11.24)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Aspergillus japonicus (Fungus)

- PDB
-
1gqg
- Quercetin 2,3-dioxygenase in complex with the inhibitor diethyldithiocarbamate
(1.7 Å)



- Catalytic CATH Domains
-
2.60.120.10
 (see all for 1gqg)
(see all for 1gqg)
- Cofactors
- Copper(2+) (1)
Enzyme Reaction (EC:1.13.11.24)
Enzyme Mechanism
Introduction
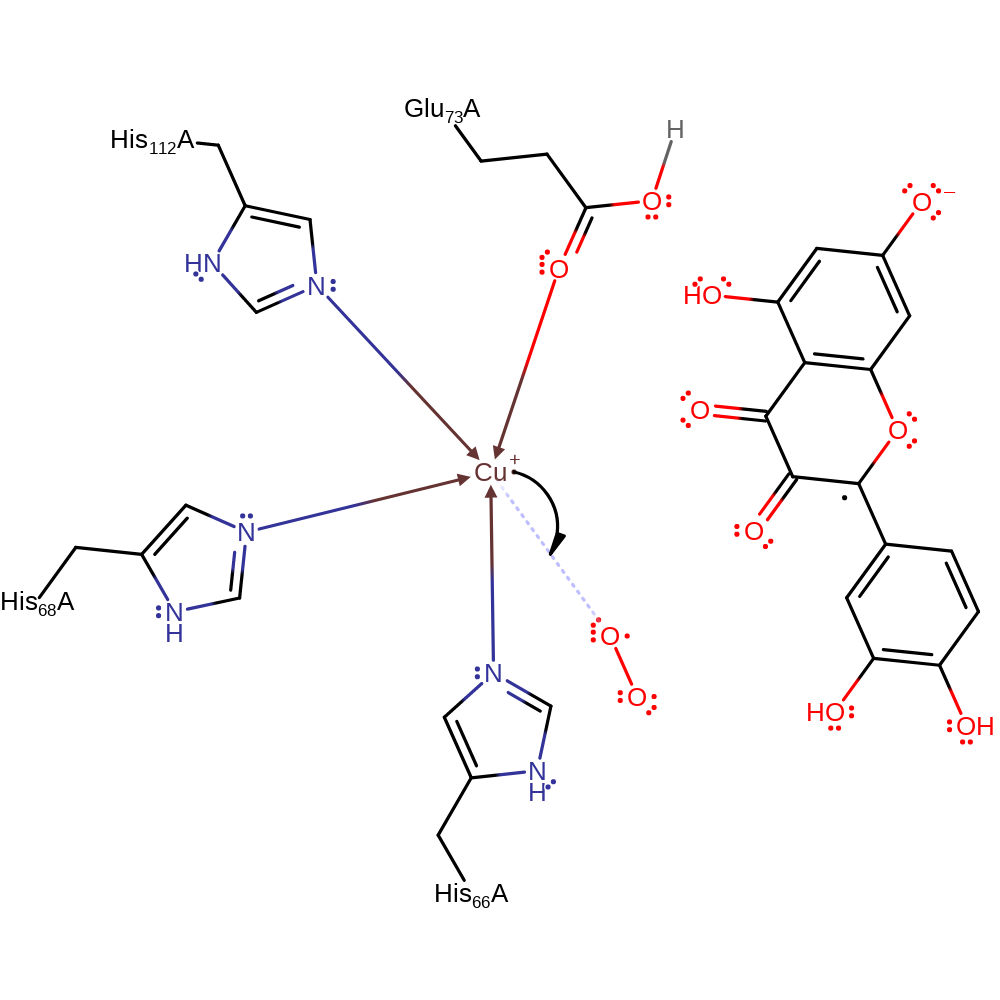
Quercetin 2,3-dioxygenase uses a Cu(II) ion to avoid problems of spin associated with reactions involving dioxygen. In the first step, the C3 hydroxyl of the substrate coordinates the Cu(II) and transfers its proton to Glu 73. Dioxygen must now bind to this complex. The ground state [Cu(II)-substrate] binds dioxygen quite poorly, and the low-lying excited state [Cu(I)-substrate(radical)] is the acceptor instead. The resulting dioxygen adduct (with three unpaired spins) is now well set up for attack by the peroxy radical on the C2 radical of the substrate, with formation of a C2-O bond.
Next, the peroxide oxygen attached to the copper forms a second C-O bond with C4, concurrent with breaking of the C4 carbonyl pi bond and bond formation between the C4 carbonyl oxygen and the copper. Glu 73 provides electrostatic stabilisation by shifting its hydrogen bond donation from the C3 substrate oxygen to the C4 substrate oxygen.
The bridging peroxide O-O bond is now cleaved, with simultaneous cleavage of the C3-C2 and C3-C4 bonds to release C3 as carbon monoxide and form a new ketone at C2 and carboxylate (still coordinated to copper) at C4. Finally, the proton stored on Glu 73 is transferred to the C4 carboxylate, and the product then leaves.
Catalytic Residues Roles
| UniProt | PDB* (1gqg) | ||
| His66, His112, His68 | His66A, His112A, His68A | The residues are bound to the Cu(II) centre. | metal ligand |
| Glu73 | Glu73A | Glu73 acts as a proton acceptor and donor in the first and last steps of the mechanism respectively. | proton acceptor, electrostatic stabiliser, proton donor |
Chemical Components
proton transfer, overall reactant used, electron transfer, radical propagation, intermediate formation, elimination (not covered by the Ingold mechanisms), overall product formed, native state of enzyme regeneratedReferences
- Fusetti F et al. (2002), Structure, 10, 259-268. Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus. PMID:11839311.
- Malkhasian AY et al. (2016), J Biomol Struct Dyn, 34, 2453-2461. Docking and DFT studies on ligand binding to Quercetin 2,3-dioxygenase. DOI:10.1080/07391102.2015.1123190. PMID:26599260.
- Fiorucci S et al. (2006), Proteins, 64, 845-850. Molecular simulations reveal a new entry site in quercetin 2,3-dioxygenase. A pathway for dioxygen? DOI:10.1002/prot.21042. PMID:16786599.
- Siegbahn PE (2004), Inorg Chem, 43, 5944-5953. Hybrid DFT Study of the Mechanism of Quercetin 2,3-Dioxygenase. DOI:10.1021/ic0498541. PMID:15360243.

Step 1. Glu73 deprotonates the hydroxyl group of the substrate which then binds to the Cu(II) through the oxygen. The substrate rearranges to form a radical intermediate in the low-lying excited state [Cu(I)-substrate(radical)].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | proton acceptor |
Chemical Components
proton transfer, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | electrostatic stabiliser |
Chemical Components
electron transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
Chemical Components
radical propagation, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components

Step 5. There is simultaneous cleavage of the peroxide O-O bond and two C-C bonds. Carbon monoxide is released.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | electrostatic stabiliser |
Chemical Components
elimination (not covered by the Ingold mechanisms), overall product formed
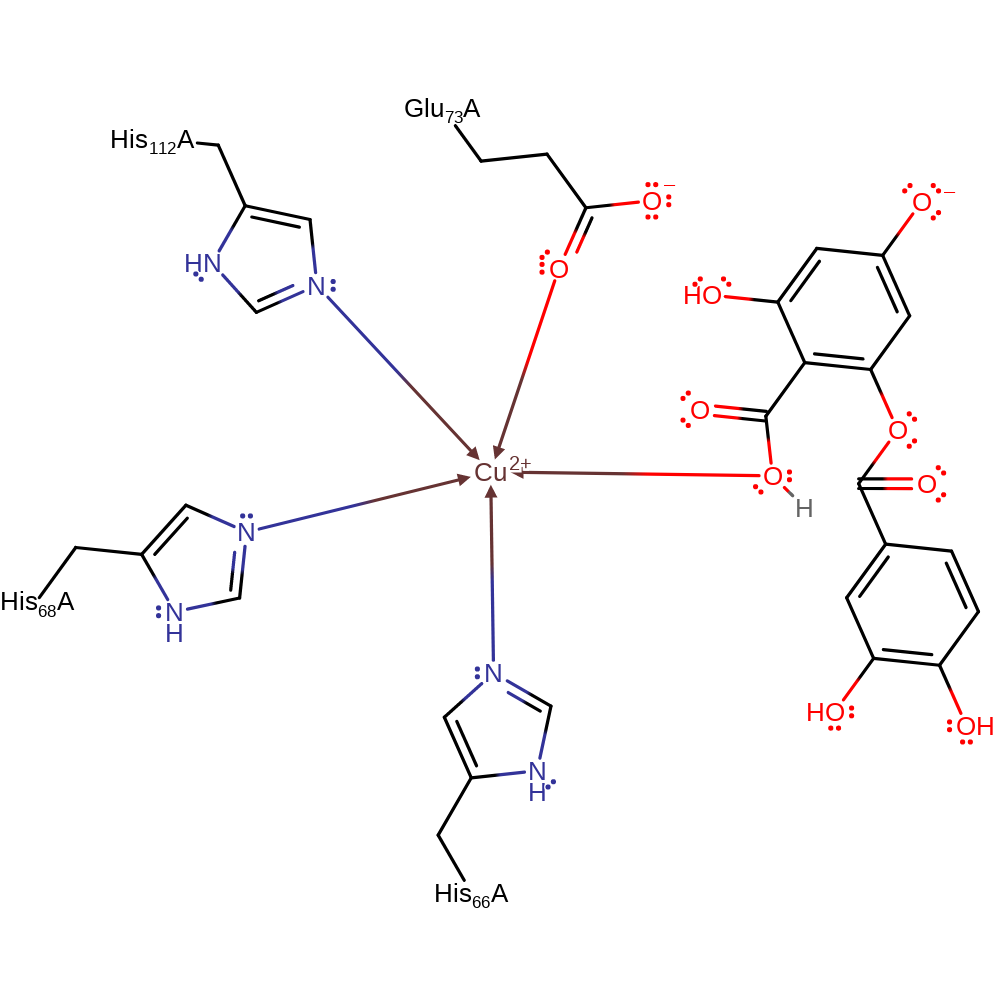
Step 6. Glu73 donates a proton to the product, completing the reaction and regenerating the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | proton donor |
Chemical Components
proton transfer, overall product formed, native state of enzyme regeneratedIntroduction
Quercetin 2,3-dioxygenase uses a Cu(II) ion to avoid problems of spin associated with reactions involving dioxygen. In the first step, the C3 hydroxyl of the substrate coordinates the Cu(II) and transfers its proton to Glu 73. Dioxygen must now bind to this complex. The ground state [Cu(II)-substrate] binds dioxygen quite poorly, and the low-lying excited state [Cu(I)-substrate(radical)] is the acceptor instead. Dioxygen attacks the substrate radical directly, with formation of a C2-O bond.
Next, the peroxide oxygen forms a second C-O bond with C4, concurrent with breaking of the C4 carbonyl pi bond and bond formation between the C4 carbonyl oxygen and the copper. Glu 73 provides electrostatic stabilisation by shifting its hydrogen bond donation from the C3 substrate oxygen to the C4 substrate oxygen.
The bridging peroxide O-O bond is now cleaved, with simultaneous cleavage of the C3-C2 and C3-C4 bonds to release C3 as carbon monoxide and form a new ketone at C2 and carboxylate (still coordinated to copper) at C4. Finally, the proton stored on Glu 73 is transferred to the C4 carboxylate, and the product then leaves.
Catalytic Residues Roles
| UniProt | PDB* (1gqg) | ||
| His66, His112, His68 | His66A, His112A, His68A | The residues are bound to the Cu ion. | metal ligand |
| Glu73 | Glu73A | Accepts proton from C3 hydroxyl of substrate. Acts as a hydrogen bond donor to the C3 and later the C4 substrate oxygens. | proton acceptor, electrostatic stabiliser, proton donor |
Chemical Components
proton transfer, coordination to a metal ion, overall reactant used, radical propagation, homolysis, rate-determining step, elimination (not covered by the Ingold mechanisms), overall product formed, native state of enzyme regeneratedReferences
- Steiner RA et al. (2002), Biochemistry, 41, 7955-7962. Functional Analysis of the Copper-Dependent Quercetin 2,3-Dioxygenase. 1. Ligand-Induced Coordination Changes Probed by X-ray Crystallography: Inhibition, Ordering Effect, and Mechanistic Insights†. DOI:10.1021/bi0159736. PMID:12069585.
- Xie H et al. (2012), Sci China Chem, 55, 1832-1841. Oxygenolysis reaction mechanism of copper-dependent quercetin 2,3-dioxygenase: A density functional theory study. DOI:10.1007/s11426-012-4729-0.
- Antonczak S et al. (2009), Phys Chem Chem Phys, 11, 1491-1501. Theoretical investigations of the role played by quercetinase enzymes upon the flavonoids oxygenolysis mechanism. DOI:10.1039/b814588a. PMID:19240925.
- Siegbahn PE (2004), Inorg Chem, 43, 5944-5953. Hybrid DFT Study of the Mechanism of Quercetin 2,3-Dioxygenase. DOI:10.1021/ic0498541. PMID:15360243.

Step 1. Glu73 deprotonates the hydroxyl group of the substrate which then binds to the Cu(II) through the oxygen. There is tautomerisation of the resulting enol, forming a radical intermediate in the low-lying excited state [Cu(I)-substrate(radical)].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu73A | electrostatic stabiliser |
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | proton acceptor |
Chemical Components
proton transfer, coordination to a metal ion, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | electrostatic stabiliser |
Chemical Components
radical propagation
Step 3. There is radical attack of the peroxide the on carbonyl carbon of the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | electrostatic stabiliser |
Chemical Components
radical propagationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
Chemical Components
homolysis, radical propagation
Step 5. There is simultaneous cleavage of the Oa–Ob and C2–C3 bonds, accompanied by the release of a CO molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
Chemical Components
rate-determining step, elimination (not covered by the Ingold mechanisms), overall product formed
Step 6. The oxyanion accepts a proton from Glu73 to form the product and regenerate the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His66A | metal ligand |
| His68A | metal ligand |
| His112A | metal ligand |
| Glu73A | proton donor |






 Download:
Download: 

 Download:
Download: