CAMP-dependent protein kinase
Cyclic adenosine 3',5'-monophosphate (cAMP) dependent protein kinase, otherwise known as protein kinase A (PKA) from Bos taurus catalyses the transfer of the gamma phosphate from ATP to a protein. This enzyme can phosphorylate a number of different proteins, one of which being the 70-kDa heat-shock protein (Hsp70). This results in the deactivation of Hsp70. Due to their key role in cellular signalling, defects in protein kinases are often associated with cancer and other diseases. This has resulted in PKA being one of the most studied protein kinases.
Atypical protein kinase C iota (aPKC iota) has 21% similarity with PKA, sharing the same catalytic residues and thought to proceed through the same reaction mechanism. aPKC iota can phosphorylate a number of different proteins, including interleukin receptor-associated kinase. aPKCs play critical roles in signalling pathways that control cell growth, differentiation and survival. Therefore they constitute targets for the development of novel therapeutics against cancer, for aPKC iota, specifically colon cancer and chronic myelogenous leukemia.
Reference Protein and Structure
- Sequences
-
P00517
 (2.7.11.11)
(2.7.11.11)
P61926
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bos taurus (Cattle)

- PDB
-
1stc
- CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE
(2.3 Å)



- Catalytic CATH Domains
-
1.10.510.10
 (see all for 1stc)
(see all for 1stc)
- Cofactors
- Magnesium(2+) (2)
Enzyme Reaction (EC:2.7.11.11)
Enzyme Mechanism
Introduction
Asp 166 activates the phosphoacceptor, serine causing it to act as a nucleophile and attack the gamma phosphorus of ATP, creating a pentavalent transition state, stabilised by Lys 168 and Thr202 (part of the catalytic triad along with Asp167). Once the gamma phosphoryl transfer has taken place, Asp 167 accepts a proton from the phosphoacceptor. Asp 167 is stabilised by a hydrogen bond to its carboxylate O delta 1 from Thr 201. The proton accepted by Asp 166 is then transferred back to phosphoserine. PKA and PKC are thought to share the same catalytic mechanism.
Catalytic Residues Roles
| UniProt | PDB* (1stc) | ||
| Asn172 | Asn171E(A) | Forms a coordinate bond with Mg2. Mg2 is important in stabilising the leaving group ADP. | metal ligand |
| Asp185 | Asp184E(A) | Forms coordinate bonds with both magnesium ions which in turn stabilise the negative charges seen throughout the reaction. | metal ligand |
| Asp167 | Asp166E(A) | Acts as a 'proton trap', activating the phosphoacceptor and accepting a proton from it once phosphoryl transfer has taken place. | activator, proton acceptor, proton donor |
| Lys169 | Lys168E(A) | Stabilises the transition state by hydrogen bonding to the beta-gamma bridging oxygen atom and also being in close proximity to the reactant serine's hydroxyl group, helping neutralise the negative charges seen in the active site throughout the reaction. | electrostatic stabiliser, polar interaction |
| Thr202 | Thr201E(A) | Positions Asp 167 and Lys 169 by forming hydrogen bonds to create a catalytic triad. | electrostatic stabiliser, polar interaction |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, overall reactant used, overall product formed, native state of enzyme regeneratedReferences
- Pérez-Gallegos A et al. (2015), Phys Chem Chem Phys, 17, 3497-3511. A QM/MM study of Kemptide phosphorylation catalyzed by protein kinase A. The role of Asp166 as a general acid/base catalyst. DOI:10.1039/c4cp03579h. PMID:25535906.
- Pérez-Gallegos A et al. (2014), J Comput Aided Mol Des, 28, 1077-1091. A QM/MM study of the associative mechanism for the phosphorylation reaction catalyzed by protein kinase A and its D166A mutant. DOI:10.1007/s10822-014-9786-3. PMID:25129483.
- Messerschmidt A et al. (2005), J Mol Biol, 352, 918-931. Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif. DOI:10.1016/j.jmb.2005.07.060. PMID:16125198.
- Yang J et al. (2004), J Mol Biol, 336, 473-487. Crystal Structure of a cAMP-dependent Protein Kinase Mutant at 1.26Å: New Insights into the Catalytic Mechanism. DOI:10.1016/j.jmb.2003.11.044. PMID:14757059.
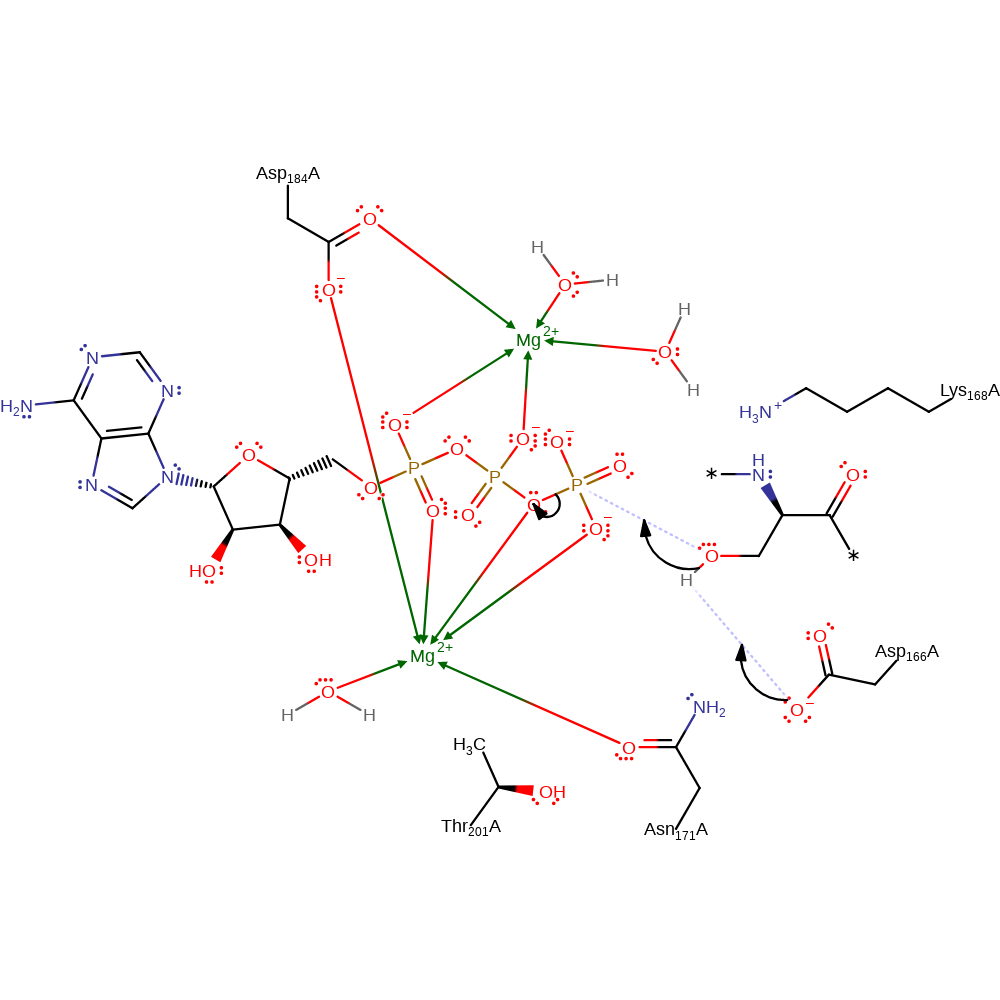
Step 1. Asp167 deprotonates a serine hydroxyl group, facilitating the nucleophilic attack on the gamma-phosphate group by the serine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr201E(A) | electrostatic stabiliser |
| Lys168E(A) | electrostatic stabiliser |
| Asp166E(A) | activator |
| Asn171E(A) | metal ligand |
| Asp184E(A) | metal ligand |
| Lys168E(A) | polar interaction |
| Thr201E(A) | polar interaction |
| Asp166E(A) | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, overall reactant used, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp184E(A) | metal ligand |
| Asn171E(A) | metal ligand |
| Asp166E(A) | proton donor |



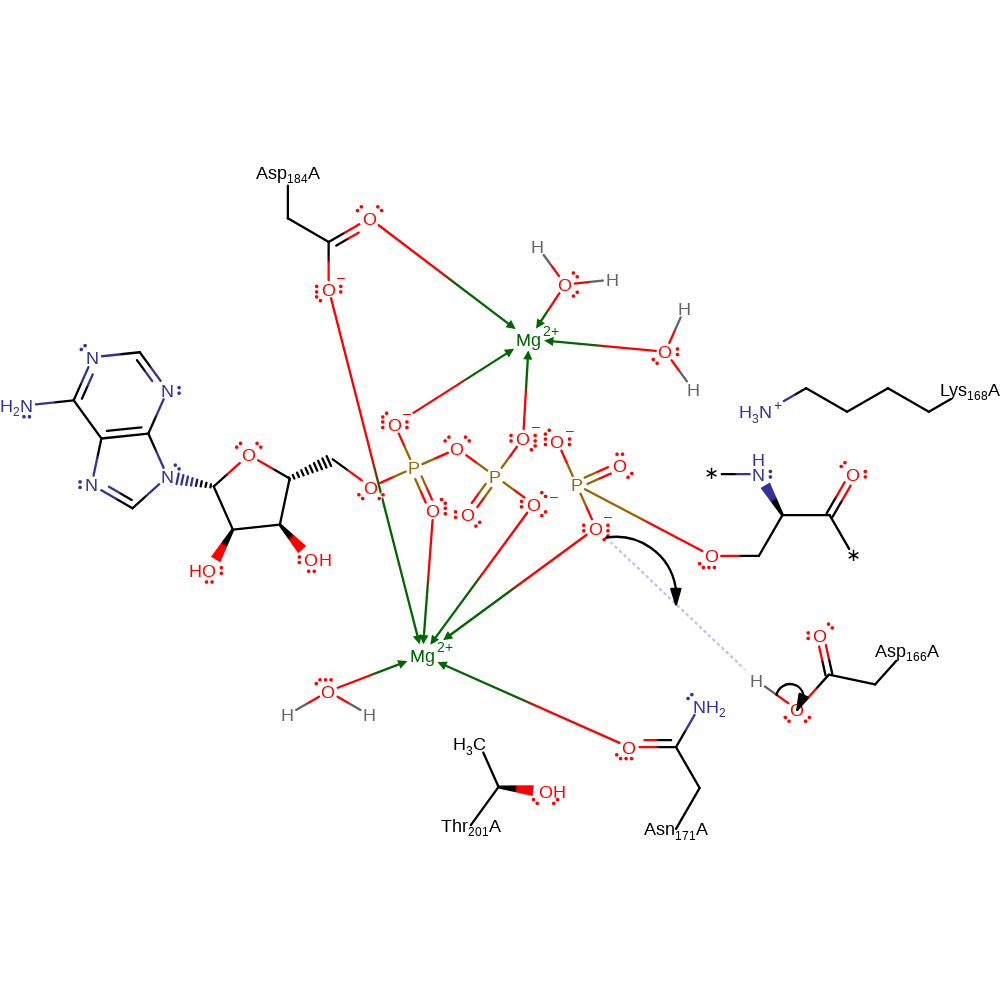
 Download:
Download: