2-hydroxymuconate-semialdehyde hydrolase
Meta-cleavage product hydrolases (MCP-hydrolase) are key enzymes involved in the microbial degradation of aromatic compounds. MCP-hydrolase CumD is a dimeric enzyme produced by Pseudomonas fluorescens IP01 and is involved in the pathway for the degradation of cumene. MCP-hydrolase enzymes produce 2-hydroxypenta-2,4-dienoate and various organic acids, depending upon the C6 substituent of the substrate. CumD prefers larger C6 substituents compared to other monoalkylbenzene hydrolases, such as TodF. Hence, when CumD acts on the meta-cleavage product of cumene, 2-hydroxypenta-2,4-dienoate and isobutyric acid are the products. The understanding of the catalytic mechanism of MCP-hydrolases is neccessary to improve biological degradation of environmental pollutant aromatic compounds, which may be carcinogenic, toxic and mutagenic.
Reference Protein and Structure
- Sequence
-
P96965

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas fluorescens (Bacteria)

- PDB
-
1uk7
- Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-butyrate
(1.7 Å)



- Catalytic CATH Domains
-
3.40.50.1820
 (see all for 1uk7)
(see all for 1uk7)
Enzyme Reaction (EC:3.7.1.9)
Enzyme Mechanism
Introduction
The active site of CumD contains the Ser103, Asp 224, His 252 catalytic triad. His 252 initially deprotonates a hydroxyl which results in the ketonization of the substrate as the keto form is more stable in the active site. His 252 is then deprotonated by the C=C so that the His can then deprotonates Ser 103 which activates it to nucleophilically attack the carbonyl bond to produce the oxyanion intermediate which is tsablised by the backbone amides of Ser 34 and Ala 103. The oxyanion initiates the first cleavage which releases the 2-oxopent-4-enoate which is then protonated His 252. His 252 then abstracts a proton from water so that it can nucleophilically attack the acyl-enzyme to form the second oxyanion intermediate which will then collapse result in the cleavage of the ester bond and the release of Ser 103 and formate.
Catalytic Residues Roles
| UniProt | PDB* (1uk7) | ||
| Ser34 (main-N), Phe104 (main-N) | Ser34A (main-N), Phe104A (main-N) | Form the oxyanion hole and stabilise the negative charge on the oxyanion | electrostatic stabiliser |
| Asp224 | Asp224A | The Asp 224 forms a hydrogen bond to the His 252 to stabilise the ion pair. | electrostatic stabiliser |
| Ser103 | Ala103A | Ser 103 is deprotonated by His 252, activating it towards nucleophilic attack of the substrate carbonyl carbon. | nucleofuge, nucleophile, proton acceptor, proton donor |
| His252 | His252A | The His 252 deprotonates an adjacent serine residue and also forms an active site ion pair with the charged serine to stabilise the transition state. | proton acceptor, proton donor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, atom stereo change, intermediate formation, overall reactant used, bimolecular electrophilic addition, bimolecular nucleophilic addition, enzyme-substrate complex formation, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regeneratedReferences
- Saku T et al. (2002), J Biosci Bioeng, 93, 568-574. Purification, characterization, and steady-state kinetics of a meta-cleavage compound hydrolase from Pseudomonas fluorescens IPO1. DOI:10.1016/s1389-1723(02)80239-0. PMID:16233251.
- Khajamohiddin S et al. (2008), Crit Rev Microbiol, 34, 13-31. Biodegradation of aromatic compounds: an overview of meta-fission product hydrolases. DOI:10.1080/10408410701683656. PMID:18259978.
- Fushinobu S et al. (2005), Biosci Biotechnol Biochem, 69, 491-498. A Series of Crystal Structures of ameta-Cleavage Product Hydrolase fromPseudomonas fluorescensIP01 (CumD) Complexed with Various Cleavage Products. DOI:10.1271/bbb.69.491. PMID:15784976.

Step 1. His252 deprotonates the hydroxyl which cause a rearrangement of double bonds which results in the ketonization of the substrate as there will be rotation around the C4-C5 bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp224A | electrostatic stabiliser |
| Ser34A (main-N) | electrostatic stabiliser |
| Phe104A (main-N) | electrostatic stabiliser |
| His252A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, atom stereo change, intermediate formation, overall reactant used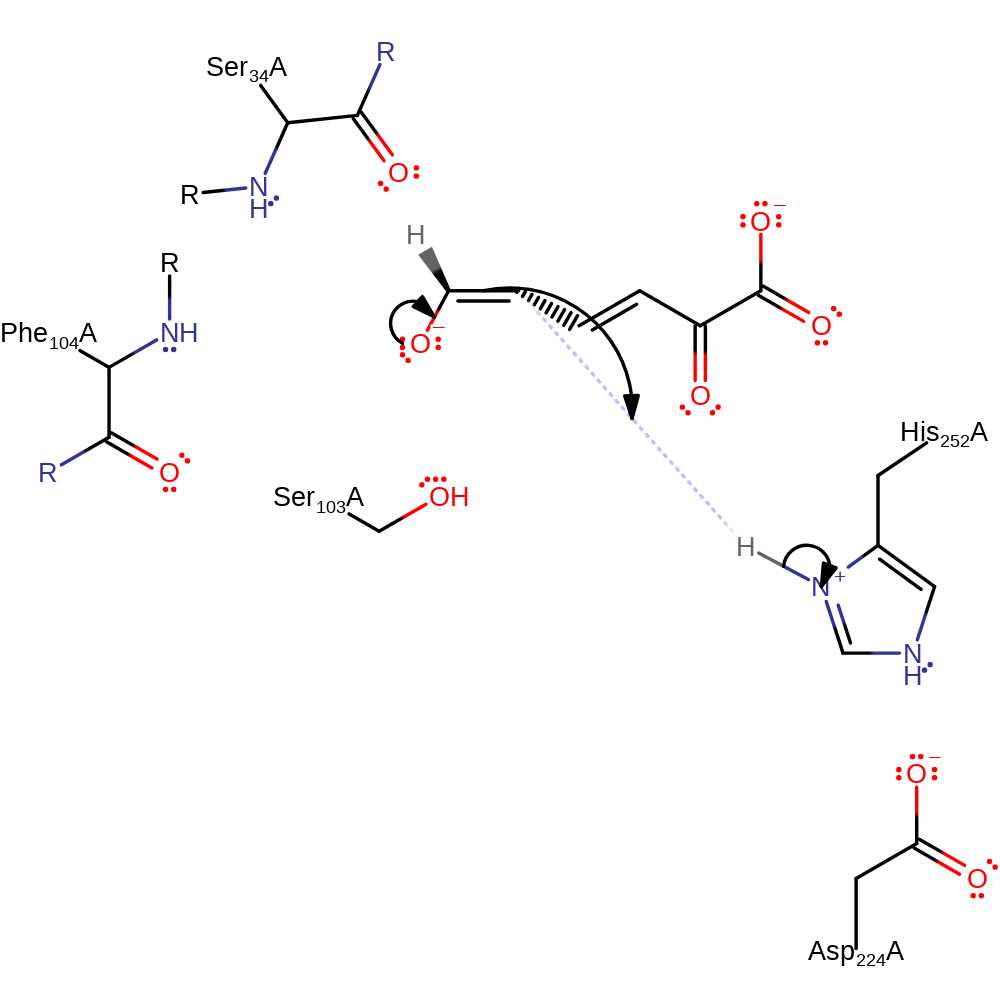
Step 2. The ketonization of the substrate generates an internal electron sink and the enolate feeds the electrons back, resulting in the C=C that is bound deprotonating His252.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser34A (main-N) | electrostatic stabiliser |
| Phe104A (main-N) | electrostatic stabiliser |
| Asp224A | electrostatic stabiliser |
| His252A | proton donor |
Chemical Components
proton transfer, intermediate formation, ingold: bimolecular electrophilic addition
Step 3. His252 deprotonates Ser103 activating it to nucleophilically attack the carbon of the carbonyl bond to produce the acyl-enzyme intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser34A (main-N) | electrostatic stabiliser |
| Phe104A (main-N) | electrostatic stabiliser |
| Asp224A | electrostatic stabiliser |
| His252A | proton acceptor |
| Ala103A | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation
Step 4. The oxyanion initiates a collapse which results in the cleavage of the intermediate which releases the 2-oxopent-4-enoate product which is protonated by His252.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser34A (main-N) | electrostatic stabiliser |
| Phe104A (main-N) | electrostatic stabiliser |
| Asp224A | electrostatic stabiliser |
| His252A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, intermediate formation, overall product formed
Step 5. His252 abstracts a proton from water, activating it to nucleophilically attack the carbon of the ester bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser34A (main-N) | electrostatic stabiliser |
| Phe104A (main-N) | electrostatic stabiliser |
| Asp224A | electrostatic stabiliser |
| His252A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 6. The oxyanion initiates an elimination which results in the cleavage of the ester bond and release of Ser103 which is then protonated by His252.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser34A (main-N) | electrostatic stabiliser |
| Phe104A (main-N) | electrostatic stabiliser |
| Asp224A | electrostatic stabiliser |
| Ala103A | nucleofuge, proton acceptor |
| His252A | proton donor |





 Download:
Download: