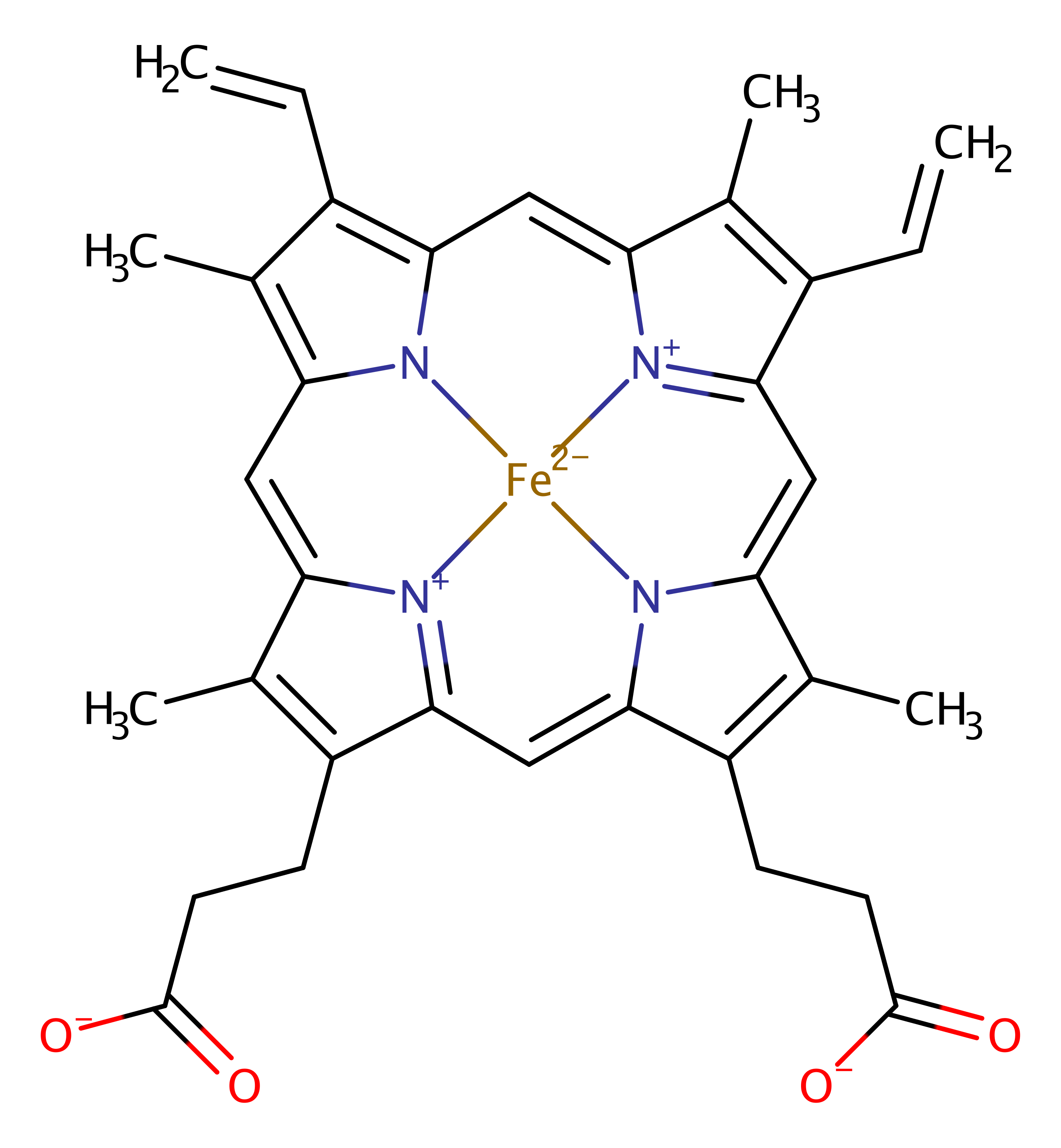
Heme oxygenase (biliverdin-producing)
Heme oxygenase (HO) catalyses the breakdown of heme to release biliverdin, CO, and free iron, and is found in mammals, plants and photosynthetic bacteria. The roles of the heme catabolism process are different in mammals and plants: whilst mammals use HO to degrade excess heme from red blood cells, plants and cyanobacteria use HO activity in order to generate the biliverdin needed for the production of light harvesting complexes. As a result, their is little sequence or structural homology between the mammalian and other forms, indicating that they may have evolved independently. However, the reaction seems to proceed via the same mechanism in both, with key active site residues being the same between the two groups. The bacterial form is described here, but the catalytic residues are the same in both the mammalian and plant forms of the enzyme.
Reference Protein and Structure
- Sequence
-
P71119
 (1.14.14.18)
(1.14.14.18)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Corynebacterium diphtheriae NCTC 13129 (Bacteria)

- PDB
-
1wnw
- D136N mutant of Heme Oxygenase from Corynebacterium diphtheriae (HmuO)
(1.7 Å)



- Catalytic CATH Domains
-
1.20.910.10
 (see all for 1wnw)
(see all for 1wnw)
Enzyme Reaction (EC:1.14.14.18)
Enzyme Mechanism
Introduction
The reaction proceeds via a series of reaction intermediates involving the heme iron in different oxidation states. In the first stage of the reaction, dioxygen binds to the iron (III) ion in heme, and accepts an electron from the surrounding environment causing heterolytic fission of the pi bond. Concomitant protonation of the dioxygen by a water molecule, activated by Asp 136, results in the formation of an Fe-OOH species. The OH group then acts as a nucleophile, attacking the alpha position of the porphyrin ring. Steric strain from Gly 135 and Gly 140 causes the destabilisation of all the other possible positions, thus stabilising the alpha meso hydroxylated intermediate. Binding of oxygen to this species liberates CO and forms Fe2+, which is then released when another molecule of dioxygen binds to the heme and cleaves the central cycle to form the biliverdin product. Thus the heme substrate acts as a cofactor in the reaction, by activating the dioxygen.
Catalytic Residues Roles
| UniProt | PDB* (1wnw) | ||
| His25 | His25B | Acts as the axial ligand for the heme iron. | metal ligand |
| Tyr53 | Tyr53B | The Tyr-58 side chain OH serves as a donor to the carboxylate of the catalytically critical Asp-140 | activator, electrostatic stabiliser |
| Gly140 (main-N), Gly135 (main-C) | Gly140B (main-N), Gly135B (main-C) | Through steric strain, destabilises hydroxy form of heme at every position except for alpa, thus stabilises this form relative to the others. | steric role |
| Arg132, Val131 (main-C) | Arg132B, Val131B (main-C) | Forms a hydrogen bonding network that helps activate the catalytic water molecule. | activator |
| Asp136 | Asn136B | Acts to activate a water molecule through electrostatic contacts, thus allowing the water molecule to act as an acid base for the reaction. | modifies pKa |
Chemical Components
References
- Matsui T et al. (2005), J Biol Chem, 280, 2981-2989. Roles of Distal Asp in Heme Oxygenase from Corynebacterium diphtheriae, HmuO: A WATER-DRIVEN OXYGEN ACTIVATION MECHANISM. DOI:10.1074/jbc.m410263200. PMID:15528205.
- Sato H et al. (2009), Biochem J, 419, 339-345. Crystal structure of rat haem oxygenase-1 in complex with ferrous verdohaem: presence of a hydrogen-bond network on the distal side. DOI:10.1042/BJ20082279. PMID:19154182.
- Lad L et al. (2003), J Mol Biol, 330, 527-538. Crystal structures of the ferric, ferrous, and ferrous-NO forms of the Asp140Ala mutant of human heme oxygenase-1: catalytic implications. PMID:12842469.
- Li Y et al. (2002), J Biol Chem, 277, 33018-33031. Solution NMR Characterization of an Unusual Distal H-bond Network in the Active Site of the Cyanide-inhibited, Human Heme Oxygenase Complex of the Symmetric Substrate, 2,4-Dimethyldeuterohemin. DOI:10.1074/jbc.m204216200. PMID:12070167.
- Sugishima M et al. (2002), J Biol Chem, 277, 45086-45090. Crystal Structure of Rat Heme Oxygenase-1 in Complex with Heme Bound to Azide. IMPLICATION FOR REGIOSPECIFIC HYDROXYLATION OF HEME AT THE alpha -MESO CARBON. DOI:10.1074/jbc.m207267200. PMID:12235152.
- Fujii H et al. (2001), J Am Chem Soc, 123, 6475-6484. A Role for Highly Conserved Carboxylate, Aspartate-140, in Oxygen Activation and Heme Degradation by Heme Oxygenase-1. DOI:10.1021/ja010490a. PMID:11439033.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn136B | modifies pKa |
| Tyr53B | electrostatic stabiliser, activator |
| Val131B (main-C) | activator |
| Arg132B | activator |
| Gly135B (main-C) | steric role |
| Gly140B (main-N) | steric role |
| His25B | metal ligand |