Glutamyl-tRNA reductase
Glutamyl-tRNA Reductase catalyses the reduction of the activated alpha-carboxyl group of glutamate from glutamyl-tRNA using NADPH to form glutamate-1-semialdehyde(GSA). GSA is then converted to 5-aminolevulenic acid, a precursor molecule for the biosynthesis of tetrapyrroles like chlorophylls, hemes and coenzyme B12.
Reference Protein and Structure
- Sequence
-
Q9UXR8
 (1.2.1.70)
(1.2.1.70)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Methanopyrus kandleri AV19 (Archaea)

- PDB
-
1gpj
- Glutamyl-tRNA Reductase from Methanopyrus kandleri
(1.95 Å)



- Catalytic CATH Domains
-
3.30.460.30
 (see all for 1gpj)
(see all for 1gpj)
Enzyme Mechanism
Introduction
Cys48 acts as a nucleophile to attack the alpha-carboxyl group of glutamate, which is activated by an ester linkage to tRNA, forming a highly reactive enzyme-localised thioester and tRNA is released. Direct hydride transfer from NADPH, facilitated by His84, which acts as a base catalyst to stabilise the protonated tetrahedral intermediate, leads to the formation of the product GSA.
Catalytic Residues Roles
| UniProt | PDB* (1gpj) | ||
| Cys48 | Ser48A | It acts as a nucleophile to attack the alpha-carboxyl group of glutamate activated by an ester linkage to tRNA, forming a highly reactive enzyme-localised thioester. | covalently attached, nucleofuge, nucleophile |
| His84 | His84A | It acts as a base catalyst to stabilise the protonated tetrahedral intermediate formed during the hydride transfer from NADPH to the enzyme-substrate complex, hence facilitating the hydride transfer. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, intermediate formation, overall reactant used, enzyme-substrate complex formation, aromatic unimolecular elimination by the conjugate base, bimolecular nucleophilic addition, hydride transfer, unimolecular elimination by the conjugate base, overall product formed, native state of enzyme regenerated, enzyme-substrate complex cleavageReferences
- Moser J et al. (1999), J Biol Chem, 274, 30679-30685. Methanopyrus kandleri Glutamyl-tRNA Reductase. DOI:10.1074/jbc.274.43.30679. PMID:10521455.
- Moser J et al. (2001), EMBO J, 20, 6583-6590. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. DOI:10.1093/emboj/20.23.6583. PMID:11726494.

Step 1. The sulfryl group of Cys48 acts as a nucleophile to attack glutamate and the tRNA residue is eliminated. An enzyme thioester intermediate is formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His84A | electrostatic stabiliser |
| Ser48A | covalently attached, nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, intermediate formation, overall reactant used, enzyme-substrate complex formation
Step 2. A hydride is transferred from NADPH to the thioester intermediate. This is promoted by His84.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His84A | electrostatic stabiliser |
| Ser48A | covalently attached |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: bimolecular nucleophilic addition, hydride transfer, intermediate formation
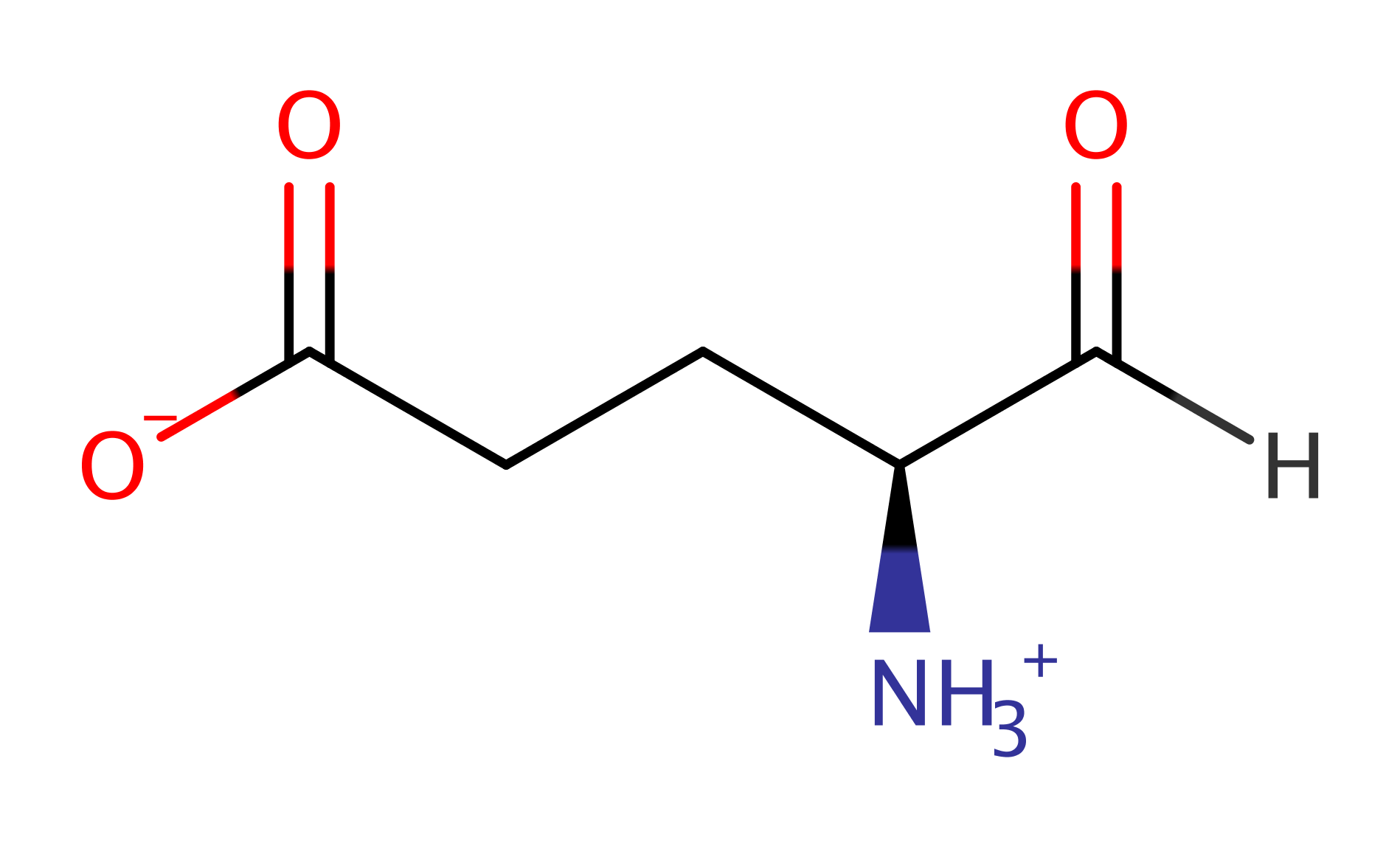
Step 3. The tetrahedral intermediate collapses, releasing Cys48 and forming the product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser48A | nucleofuge |





 Download:
Download: