Adenine DNA glycosylase
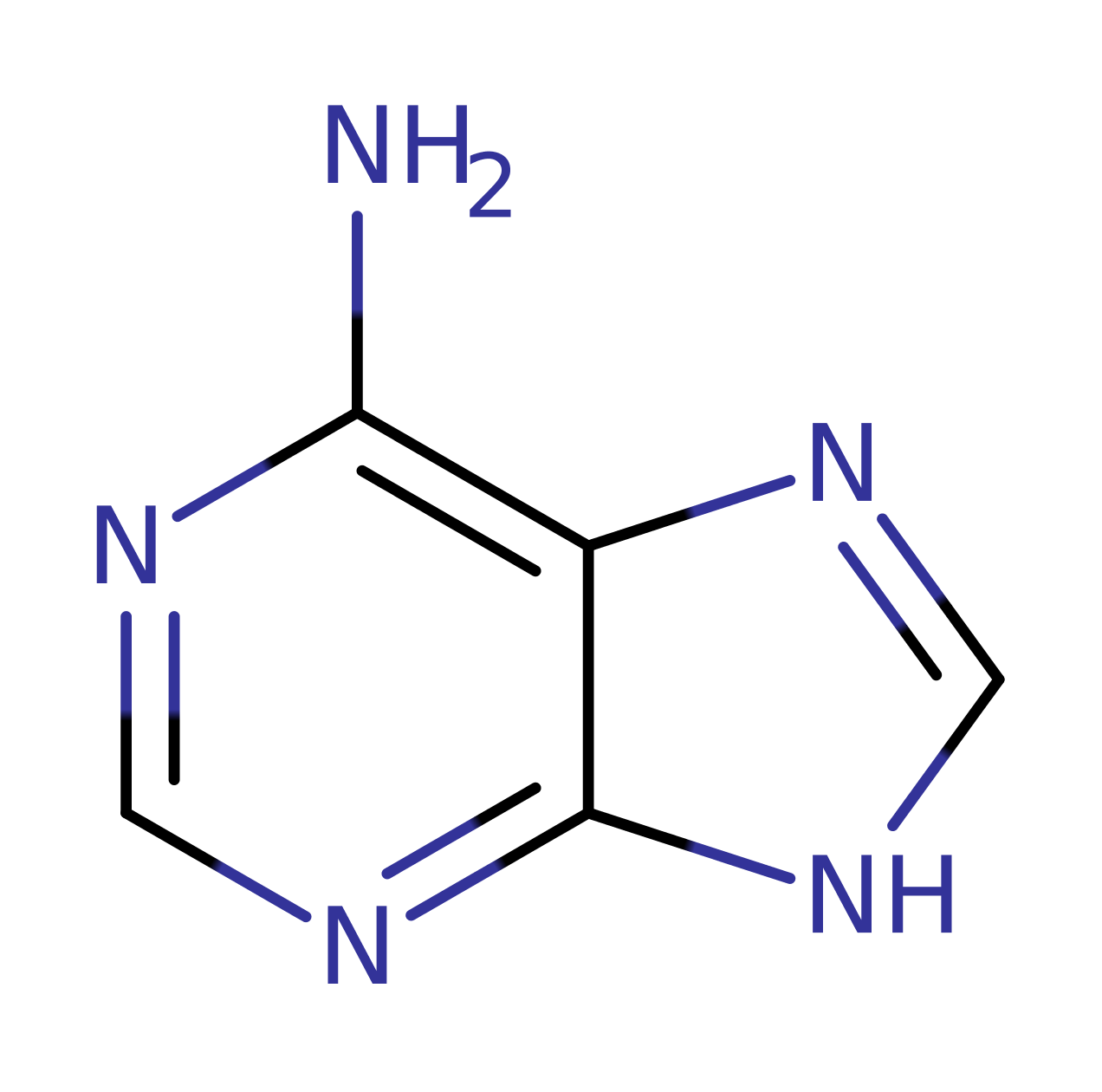
Oxidative DNA damage is caused by reactive oxygen species that are generated during aerobic respiration and immune response. The oxidative lesions in DNA caused are repaired by the base excision repair pathway. Hydroxyl radicals rapidly react with guanine C8 producing a steady state level of mutagenic 8-oxoguanines in cells. It also creates 8-oxo-GTP that, like 8-oxoguanines in DNA template, is often mispaired with adenine by replicative polymerases, creating A-T to C-G and G-C to T-A transversion mutations.
DNA glycosylase MutY recognises the mutational intermediate 8-oxoguanine-adenine mispair and excises adenine from the mispair. This creates an abasic site that is then processed by the multi-enzyme base excision repair pathway. The sequence of MutY is conserved from bacteria to humans suggesting its fundamental importance in DNA repair. Mutations of MutY increases transversion frequencies and hence cancer susceptibility in human. MutY belongs to the base excision repair glycosylases superfamily which includes both pure DNA glycosylases and DNA glycosylase/lyases. The most conserved structural element in this family is the Helix-hairpin-Helix (HhH) motif. A revised MutY reaction mechanism was published in 2016, which involves a covalent intermediate and two oxocarbenium ion-like transition states. There is sufficient structural, mutagenesis and biochemical date to support the proposed mechanism described.
Reference Protein and Structure
- Sequence
-
P83847
 (3.2.2.31)
(3.2.2.31)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Geobacillus stearothermophilus (Bacteria)

- PDB
-
6u7t
- MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with d(8-oxo-G)
(2.0 Å)



- Catalytic CATH Domains
- (see all for 6u7t)
- Cofactors
- Water (1)
Enzyme Reaction (EC:3.2.2.31)
Enzyme Mechanism
Introduction
MutY has three key catalytic residues involved in the proposed double displacement reaction consisting of base excision of adenine and hydrolysis steps with two oxocarbenium ion-like transition states. In the reaction mechanism described, the adenine would have been flipped out of a DNA polymer if it is mis-paired with 8-oxoguanine by the enzyme. The reaction mechanism begins with adenine isomerising from 2'-endo to 3'-exo conformation, enabling Glu43 to protonate adenine via a water molecule and cause hydrolytic cleavage of the N-glycosydic bond between adenine and the deoxyribose sugar in an Sn1 mechanism. Asp144 then forms a covalent intermediate with the deoxyribose sugar which is then subject to nucleophilic attack by an Glu43 activated water molecule. This results in an AP site where the adenine has been excised from the sugar-phosphate backbone in polymeric DNA.
Catalytic Residues Roles
| UniProt | PDB* (6u7t) | ||
| Glu43 | Glu43A | Glu43 is a base catalyst involved in the deprotonation of both water molecules involved in the base excision and hydrolysis reactions. | activator, proton acceptor, proton donor |
| Tyr126 | Tyr126A | Tyr126 serves to stabilise the two oxocarbenium ion-like transition states formed in the steps of base excision and hydrolysis and hydrogen bonds with Glu43. | electrostatic stabiliser |
| Asp144 | Asp144A | Asp144 acts as a nucleophile to attack the subsequent oxocarbenium ion generated after base excision to form a covalent intermediate. | covalently attached, nucleofuge, nucleophile |
Chemical Components
proton transfer, proton relay, overall reactant used, intermediate formation, elimination (not covered by the Ingold mechanisms), heterolysis, overall product formed, bimolecular nucleophilic addition, enzyme-substrate complex formation, bimolecular nucleophilic substitution, native state of enzyme regenerated, intermediate terminated, enzyme-substrate complex cleavageReferences
- Woods RD et al. (2016), Nucleic Acids Res, 44, 801-810. Structure and stereochemistry of the base excision repair glycosylase MutY reveal a mechanism similar to retaining glycosidases. DOI:10.1093/nar/gkv1469. PMID:26673696.
- McCann JA et al. (2008), J Am Chem Soc, 130, 5789-5797. Transition-state analysis of the DNA repair enzyme MutY. DOI:10.1021/ja711363s. PMID:18393424.
- Guan Y et al. (1998), Nat Struct Biol, 5, 1058-1064. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. DOI:10.1038/4168. PMID:9846876.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr126A | electrostatic stabiliser |
| Glu43A | proton donor |
Chemical Components
proton transfer, proton relay, overall reactant used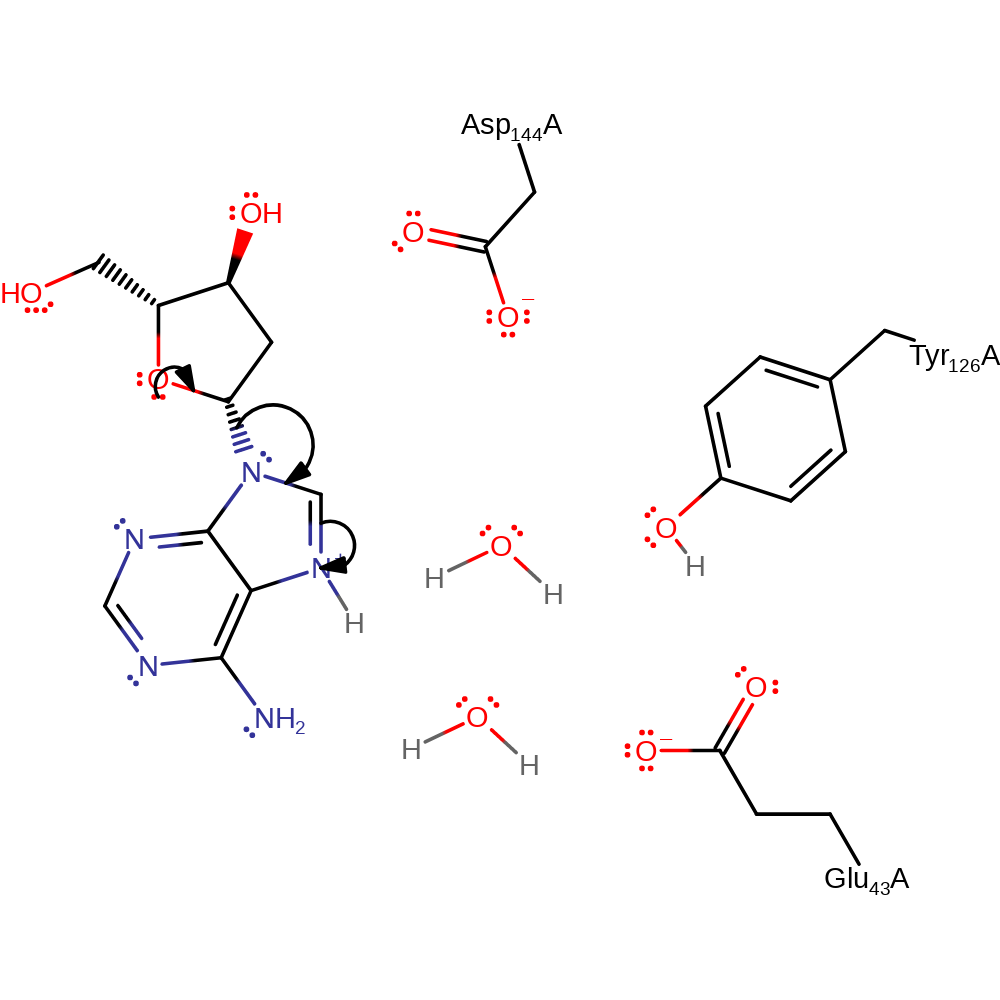
Step 2. Following adenine protonation, the N-glycosyl bond is broken to form an oxocarbenium ion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr126A | electrostatic stabiliser |
Chemical Components
intermediate formation, elimination (not covered by the Ingold mechanisms), heterolysis, overall product formed
Step 3. Asp144 attacks the oxocarbenium ion to form a covalent intermediate, enabling time for adenine to leave the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr126A | electrostatic stabiliser |
| Asp144A | covalently attached, nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, enzyme-substrate complex formation
Step 4. Another water molecule is activated by Glu43 abstracting a proton which then acts as a nucleophile and attacks the carbon of the C-O bond to Asp144. This results in the formation of 2-deoxy-D-ribofuranose and the catalytic residues are now ready to excise another adenine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu43A | activator |
| Tyr126A | electrostatic stabiliser |
| Glu43A | proton acceptor |
| Asp144A | nucleofuge |



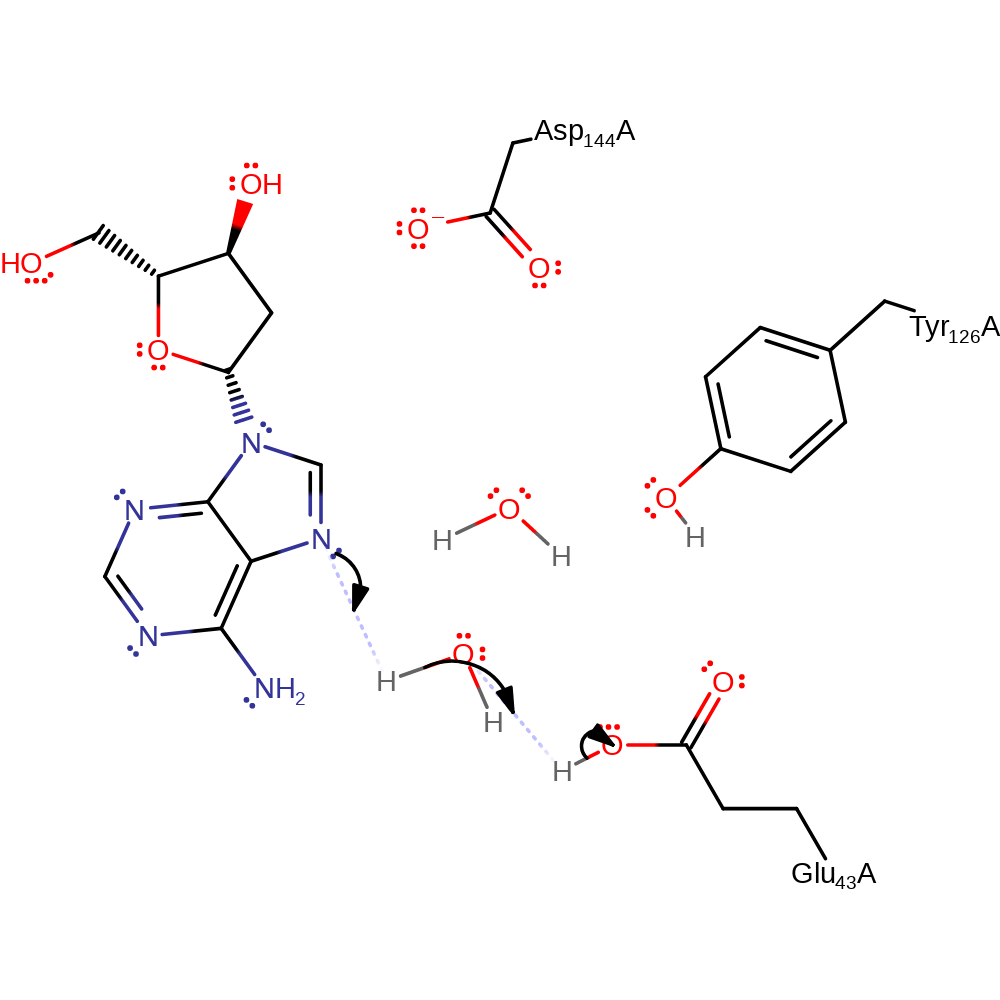
 Download:
Download: