Caspase-3
Caspase-3 from Homo sapiens is a member of a family of intracellular cysteine proteases that cleave substrates specifically at an aspartic acid residue. Caspases share a high degree of homology, not only within caspases from the same species but also across various species. Caspases control the proteolytic cascade central to the initiation and regulation of apoptosis. Caspase-3 is known as the executioner protease. The inactive form of caspase-3 is procaspase-3. This is activated when the procaspase is cleaved at an aspartate residue in the subunit linker, followed by one or more cleavages that remove the pro-domain.
Reference Protein and Structure
- Sequence
-
P42574
 (3.4.22.56)
(3.4.22.56)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1pau
- Crystal structure of the complex of apopain with the tetrapeptide aldehyde inhibitor AC-DEVD-CHO
(2.5 Å)



- Catalytic CATH Domains
-
3.40.50.1460
 (see all for 1pau)
(see all for 1pau)
Enzyme Mechanism
Introduction
Cys163 and His121 are thought to exist as a thiolate/imidazolium ion-pair. Upon binding the substrate, protonated His121 deprotonates Cys163. This enables Cys163 to perform nucleophilic attack upon the carbonyl carbon. The tetrahedral oxyanion intermediate is stabilised by the hydrogen bonding to the backbone amide group of Gly122 and Cys163 the oxyanion initiates an elimination resulting in the cleavage of the scissille bond and His121 then protonates the amino group of the N-terminal product. His121 can now deprotonate water to activate it so that it can nucleophilically attack the carbon of the thioester bond. The oxyanion formed will initiate another elimantion and this results in the breaking if the thioester bond and the release of Cys163. Cys163 can then accept a proton from His121 to regenerate the enzyme to its native state.
Catalytic Residues Roles
| UniProt | PDB* (1pau) | ||
| Thr62, Ser63 | Thr177(34)A, Ser178(35)A | Stabilise His121 through Hydrogen bonding making it more basic so more willing to accept a proton. | electrostatic stabiliser |
| Cys163 | Cys285(135)A | Cys 163 protonates His 121, causing it to become cationic. Cys 163 performs nucleophilic attack upon the activated carbonyl carbon. | nucleofuge, nucleophile, proton acceptor, proton donor |
| Cys163 (main-N), Gly122 (main-N) | Cys285(135)A (main-N), Gly238(94)A (main-N) | Gly122 and Cys163 stabilises the tetrahedral transition state through hydrogen bonding with their backbone amide group. | electrostatic stabiliser |
| His121 | His237(93)A | Acts as a general acid/base by accepting protons from Cys163 and water to activate them for nucleophilic attack and and protonates amino group to prevent pepide reformation. | proton acceptor, electrostatic stabiliser, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, intermediate formation, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Chéreau D et al. (2003), Biochemistry, 42, 4151-4160. Structural and Functional Analysis of Caspase Active Sites. DOI:10.1021/bi020593l. PMID:12680769.
- Sulpizi M et al. (2003), Proteins, 52, 212-224. Reaction mechanism of caspases: insights from QM/MM Car-Parrinello simulations. DOI:10.1002/prot.10275. PMID:12833545.
- Brady KD et al. (1999), Bioorg Med Chem, 7, 621-631. A catalytic mechanism for caspase-1 and for bimodal inhibition of caspase-1 by activated aspartic ketones. DOI:10.1016/s0968-0896(99)00009-7. PMID:10353641.
- Stennicke HR et al. (1999), Cell Death Differ, 6, 1054-1059. Catalytic properties of the caspases. DOI:10.1038/sj.cdd.4400599. PMID:10578173.

Step 1. His121 deprotonates Cys163 thus activating it to nucleophilically attack the carbon of the carbonyl of the peptide bond which produces the oxyanion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His237(93)A | electrostatic stabiliser |
| Gly238(94)A (main-N) | electrostatic stabiliser |
| Thr177(34)A | electrostatic stabiliser |
| Ser178(35)A | electrostatic stabiliser |
| Cys285(135)A | nucleophile, proton donor |
| His237(93)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation
Step 2. The oxyanion initiates an elimantion resulting in the cleavage of the peptide bond. The N-terminal product accepts a proton from His121.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr177(34)A | electrostatic stabiliser |
| Ser178(35)A | electrostatic stabiliser |
| Gly238(94)A (main-N) | electrostatic stabiliser |
| Cys285(135)A (main-N) | electrostatic stabiliser |
| His237(93)A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, overall product formed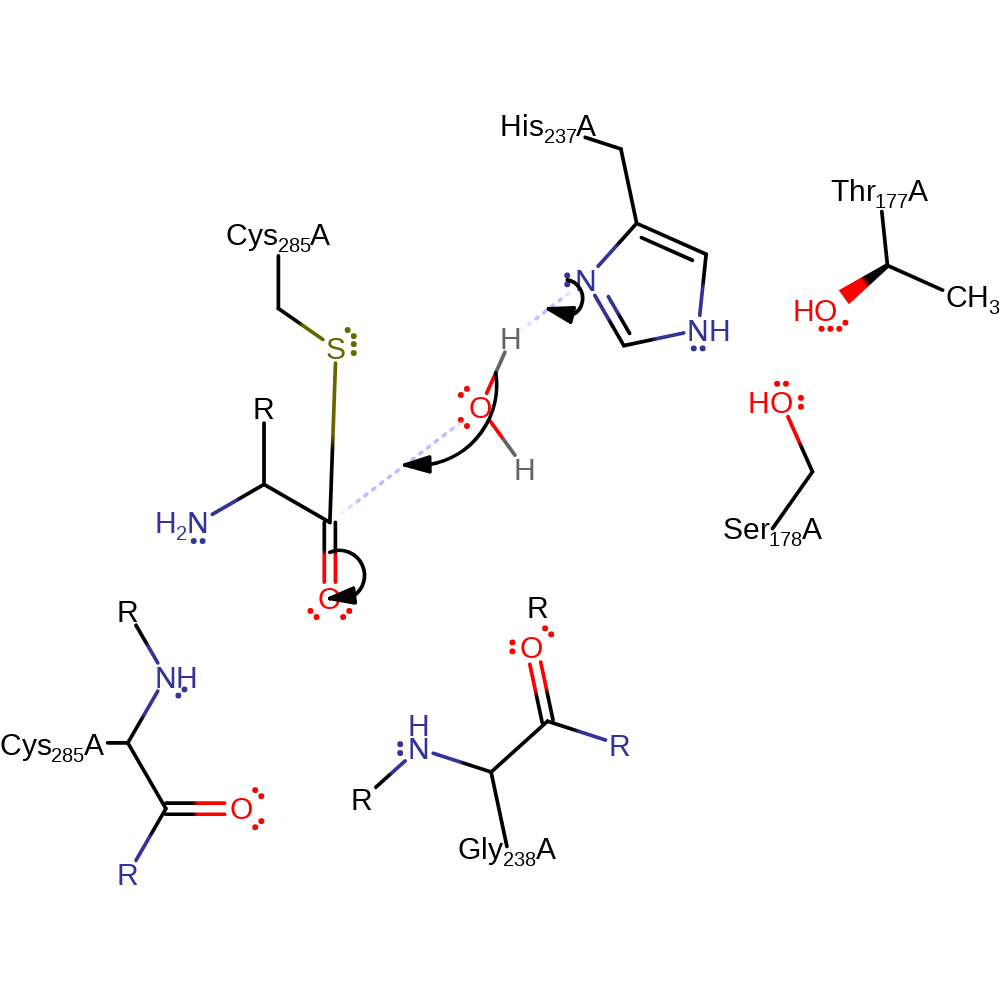
Step 3. His121 deprotonates water activating it so that it can attack the carbon of the thioester bond in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr177(34)A | electrostatic stabiliser |
| Ser178(35)A | electrostatic stabiliser |
| Gly238(94)A (main-N) | electrostatic stabiliser |
| Cys285(135)A (main-N) | electrostatic stabiliser |
| His237(93)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 4. The oxyanion initiates an elimantion resulting in the cleavage of the thioester bond and Sulfur of Cys163 can then accept a proton from His121.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr177(34)A | electrostatic stabiliser |
| Ser178(35)A | electrostatic stabiliser |
| Gly238(94)A (main-N) | electrostatic stabiliser |
| Cys285(135)A (main-N) | electrostatic stabiliser |
| Cys285(135)A | nucleofuge |
| His237(93)A | proton donor |
| Cys285(135)A | proton acceptor |



 Download:
Download: