Glutamine-fructose-6-phosphate transaminase (isomerizing)
Glucosamine-fructose-6-phosphate aminotransferase (isomerizing) also known as Glucosamine-6-phosphate synthase (GlmS), catalyses the first reaction in hexamine biosynthesis. It belongs to an F-type group of glutamine-dependent amidotransferase family of enzymes, which utilise the glutamine amide nitrogen in the biosynthesis of phosphoribosylamine, glutamate or asparagine.
The hexosamine biosynthetic pathway starts from D fructose-6-phosphate (Fru6P)2 which is produced from glucose via the glycolysis pathway. The Fru6P is converted into D -glucosamine-6-phosphate (GlcN6P) by the rate-limiting enzyme glucosamine-6-phosphate synthase (GlcN6P synthase). This is the sole biosynthetic route to GlcN6P known to date.
The reaction is practically irreversible and the reaction takes place over two structural domains, an N-terminal glutaminase domain, which hydrolyses glutamine to glutamate and ammonia (residues 1-240), and a C-terminal isomerase domain (residues 241-608), which catalyses the ketose-aldose isomerisation and utilises the nitrogen for synthesis of GlcN-6P.
The isomerase domain is responsible for two activities of GlmS, the conversion of Fru-6P into GlcN-6P in the presence of glutamine (the synthase activity), and the isomerisation of Fru-6P into Glc-6P (the phosphoglucose isomerase - like activity) in the absence of glutamine.
The product of the reaction with fructose 6-phosphate, glucosamine 6-phosphate, undergoes transformation leading towards formation of uridine diphospho-N-acetylglucosamine - which is a precursor to all amino sugar-containing macromolecules. Much interest has been shown in this enzyme that is believed to have important implications in antibacterial/antifungal therapy and diabetes treatment.
Reference Protein and Structure
- Sequence
-
P17169
 (2.6.1.16)
(2.6.1.16)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1jxa
- GLUCOSAMINE 6-PHOSPHATE SYNTHASE WITH GLUCOSE 6-PHOSPHATE
(3.1 Å)



- Catalytic CATH Domains
-
3.60.20.10
 3.40.50.10490
3.40.50.10490  (see all for 1jxa)
(see all for 1jxa)
- Cofactors
- Water (1)
Enzyme Reaction (EC:2.6.1.16)
Enzyme Mechanism
- Summary
- Step 1
- Step 2
- Step 3
- Step 4
- Step 5
- Step 6
- Step 7
- Step 8
- Step 9
- Step 10
- Step 11
- Step 12
- Step 13
- Step 14
- Products
- All Steps
Introduction
In the GAT domain deprotonated Cys1 attacks the amide carbonyl of L-glutamine forming a tetrahedral intermediate. Gly99 and Asn98 stabilise negative charge by formation of an oxyanion hole via hydrogen bonding. Subsequent collapse leads to release of ammonia, which is then transferred to the isomerase domain via a channel. In the GAT domain a deprotonated water molecule then attacks the carbonyl group to form a second tetrahedral intermediate with collapse leading to release of Cys1. Note during the reaction in GAT domain the base that deprotonates water is the N-terminus of Cys1.
In the isomerase domain His504 deprotonates the hydroxyl group of C2, which leads to ring opening of Fru6P. Then Lys603 forms an imine linkage with Fru6P via nucleophilic attack. Ammonia formed in the GAT domain then attacks this linkage leading to nucleophilic substituion. Glu488 deprotonates C1 with concommitant double bond rearrangement that deprotonates Lys485. Lys485 then deprotonates C1-OH with concommitant double bond rearrangement and deprotonation of Glu488. Finally His504 deprotonates C5 hydroxyl,the resulting oxyanion initiates a nucleophilic attack on the C1 carbonyl carbon in an addition reaction with concomitant deprotonation of His504B leading to formation of the GlcN6P product.
Catalytic Residues Roles
| UniProt | PDB* (1jxa) | ||
| Glu482 | Glu481A | Stabilises and activates the catalytic lysine (Lys485). | hydrogen bond acceptor, electrostatic stabiliser |
| Cys2 | Cys1A | Acts as the catalytic nucleophile in the deamination reaction. It is activated by its own main chain amide (which is in the form of the N-terminus of the protein). | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor |
| Trp75 (main-N), Arg27 (main-C) | Trp74A (main-N), Arg26A (main-C) | Stabilise the formation of protonated N-terminus of Cys1 via hydrogen bonding. | hydrogen bond donor, electrostatic stabiliser |
| Cys2 (N-term) | Cys1A (N-term) | N-terminus acts as a base to deprotonate water, which in turn deprotonates the thiol proton to form a nucleophile. When ammonia leaves it picks up a proton from the N-terminus. The N-terminus then deprotonates another molecule of water to form a hydroxide nucleophile. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu489 | Glu488A | Acts as a general acid/base in the deprotonation of C1 to form the isomer. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Lys604 | Lys603A | Acts as a nucleophile to form imine linkage with carbonyl group. | covalently attached, hydrogen bond donor, nucleofuge, proton acceptor, proton donor, nucleophile, electron pair acceptor, electron pair donor |
| Lys486 | Lys485A | Acts as a general acid/base in the isomerisation of Fru6p to Glc6P. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Gly100 (main-N), Asn99 | Gly99A (main-N), Asn98A | Forms a hydrogen bond to the negatively charged oxygen to stabilise its formation, via an oxyanion hole. | hydrogen bond donor, electrostatic stabiliser |
| His505 | His504A(AA) | Acts as a general base to initiate decyclisation of Fru6P. Then protonates in the final step to form Glc6P. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton relay, unimolecular elimination by the conjugate base, deamination, enzyme-substrate complex cleavage, intermediate collapse, overall product formed, native state of enzyme regenerated, intermediate terminated, bimolecular elimination, decyclisation, dehydration, schiff base formed, intramolecular elimination, assisted tautomerisation (not keto-enol), assisted keto-enol tautomerisation, intramolecular nucleophilic addition, cyclisationReferences
- Durand P et al. (2008), Arch Biochem Biophys, 474, 302-317. Highlights of glucosamine-6P synthase catalysis. DOI:10.1016/j.abb.2008.01.026. PMID:18279655.
- Chennu MM et al. (2015), Bioinformation, 11, 525-528. Molecular docking based screening of G6PS with 1, 5 Benzothiazepine derivates for a potential inhibitor. DOI:10.6026/97320630011525. PMID:26770025.
- Jędrzejczak R et al. (2012), Chembiochem, 13, 85-96. Inactivation of Glucosamine-6-Phosphate Synthase by N3-Oxoacyl Derivatives of L-2,3-Diaminopropanoic Acid. DOI:10.1002/cbic.201100587. PMID:22125025.
- Mouilleron S et al. (2011), Arch Biochem Biophys, 505, 1-12. Dynamics of glucosamine-6-phosphate synthase catalysis. DOI:10.1016/j.abb.2010.08.008. PMID:20709015.
- Floquet N et al. (2009), J Mol Biol, 385, 653-664. Collective motions in Glucosamine-6-phosphate Synthase: Influence of Ligand Binding and role in Ammonia Channelling and Opening of the Fructose-6-Phosphate Binding Site. DOI:10.1016/j.jmb.2008.10.032. PMID:18976669.
- Mouilleron S et al. (2008), J Mol Biol, 377, 1174-1185. Ordering of C-terminal Loop and Glutaminase Domains of Glucosamine-6-Phosphate Synthase Promotes Sugar Ring Opening and Formation of the Ammonia Channel. DOI:10.1016/j.jmb.2008.01.077. PMID:18295797.
- Mouilleron S et al. (2007), Protein Sci, 16, 485-493. Domain motions of glucosamine-6P synthase: Comparison of the anisotropic displacements in the crystals and the catalytic hinge-bending rotation. DOI:10.1110/ps.062598107. PMID:17322533.
- Mouilleron S et al. (2006), J Biol Chem, 281, 4404-4412. Glutamine Binding Opens the Ammonia Channel and Activates Glucosamine-6P Synthase. DOI:10.1074/jbc.m511689200. PMID:16339762.
- (2002), Nat Prod Rep, 19, 60-69. From Lobry de Bruyn to enzyme-catalyzed ammonia channelling: molecular studies of D-glucosamine-6P synthase. DOI:10.1039/b103713g.
- Milewski S (2002), Biochim Biophys Acta, 1597, 173-192. Glucosamine-6-phosphate synthase—the multi-facets enzyme. DOI:10.1016/s0167-4838(02)00318-7. PMID:12044898.
- Teplyakov A et al. (2001), J Mol Biol, 313, 1093-1102. Channeling of ammonia in glucosamine-6-phosphate synthase. DOI:10.1006/jmbi.2001.5094. PMID:11700065.
- Oinonen C et al. (2000), Protein Sci, 9, 2329-2337. Structural comparison of Ntn-hydrolases. DOI:10.1110/ps.9.12.2329. PMID:11206054.
- Bearne SL et al. (2000), J Biol Chem, 275, 135-140. Inhibition of Escherichia coliGlucosamine-6-phosphate Synthase by Reactive Intermediate Analogues: THE ROLE OF THE 2-AMINO FUNCTION IN CATALYSIS. DOI:10.1074/jbc.275.1.135.
- Teplyakov A et al. (1999), Protein Sci, 8, 596-602. The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase. DOI:10.1110/ps.8.3.596. PMID:10091662.
- Teplyakov A et al. (1998), Structure, 6, 1047-1055. Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: evidence from a 1.6 A crystal structure of the isomerase domain. PMID:9739095.
- Isupov MN et al. (1996), Structure, 4, 801-810. Substrate binding is required for assembly of the active conformation of the catalytic site in Ntn amidotransferases: evidence from the 1.8 å crystal structure of the glutaminase domain of glucosamine 6-phosphate synthase. DOI:10.1016/s0969-2126(96)00087-1. PMID:8805567.
- Bearne SL et al. (1995), Biochemistry, 34, 11515-11520. Glutamate gamma-semialdehyde as a natural transition state analogue inhibitor of Escherichia coli glucosamine-6-phosphate synthase. PMID:7547881.
- Obmolova G et al. (1994), J Mol Biol, 242, 703-705. Crystallization and Preliminary X-ray Analysis of the Two Domains of Glucosamine-6-phosphate Synthase from Escherichia coli. DOI:10.1006/jmbi.1994.1619. PMID:7932726.
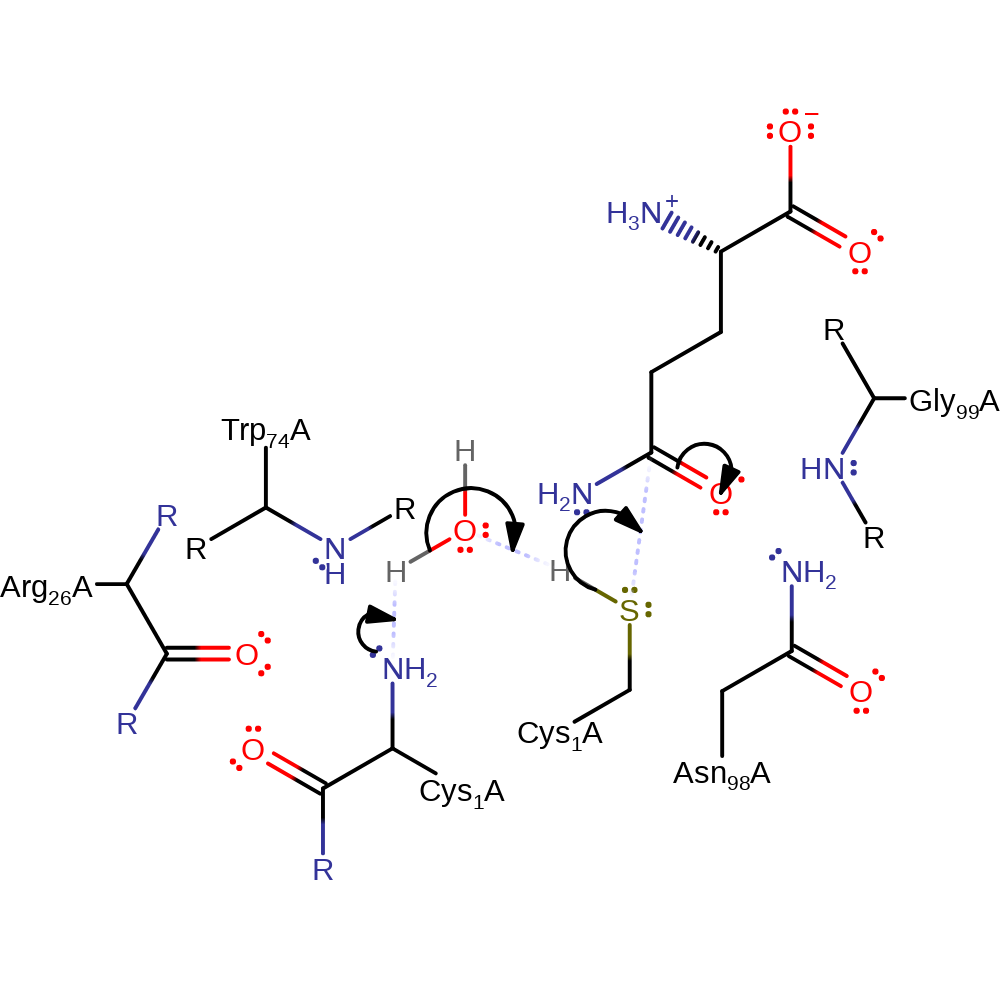
Step 1. The N-terminus of Cys1 deprotonates water, which then deprotonates the thiol group of Cys1, initiating a nucleophilic attack on the amide carbon in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn98A | hydrogen bond donor, electrostatic stabiliser |
| Gly99A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A (N-term) | hydrogen bond acceptor |
| Arg26A (main-C) | hydrogen bond acceptor, activator |
| Trp74A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A | proton donor, nucleophile |
| Cys1A (N-term) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton relay
Step 2. The oxyanion initiates an elimination that cleaves ammonia from the bound L-glutamine substrate. Ammonia deprotonates water, which deprotonates the N-terminus of Cys1.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn98A | hydrogen bond donor, electrostatic stabiliser |
| Gly99A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A (N-term) | hydrogen bond donor |
| Arg26A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Trp74A (main-N) | hydrogen bond donor |
| Cys1A | covalently attached |
| Cys1A (N-term) | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, proton relay, deamination, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation
Step 3. The N-terminus of Cys1 deprotonates water, which initiates a nucleophilic attack on the carbonyl carbon of the covalently bound intermediate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn98A | hydrogen bond donor, electrostatic stabiliser |
| Gly99A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Arg26A (main-C) | hydrogen bond acceptor, activator |
| Trp74A (main-N) | hydrogen bond donor |
| Cys1A (N-term) | hydrogen bond acceptor |
| Cys1A | covalently attached |
| Cys1A (N-term) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, proton relay, enzyme-substrate complex formation, intermediate formation
Step 4. The oxyanion initiates an elimination that cleaves the C-S bond, the thiolate of Cys1 deprotonates water, which deprotonates the N-terminus of Cys1
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn98A | hydrogen bond donor, electrostatic stabiliser |
| Gly99A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A (N-term) | hydrogen bond acceptor, hydrogen bond donor |
| Arg26A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Trp74A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A | nucleofuge, proton acceptor |
| Cys1A (N-term) | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, overall product formed, native state of enzyme regenerated, enzyme-substrate complex cleavage, proton relay, intermediate terminated, intermediate collapse
Step 5. His504B deprotonates the hydroxyl group of C2 carbon in a decyclisation step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His504A(AA) | hydrogen bond acceptor |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| His504A(AA) | proton acceptor |
Chemical Components
ingold: bimolecular elimination, overall reactant used, decyclisation, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His504A(AA) | hydrogen bond donor |
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Lys603A | hydrogen bond donor |
| His504A(AA) | proton donor |
Chemical Components
proton transfer, intermediate formation
Step 7. The C2 carbonyl oxygen deprotonates Lys603, initiating a nucleophilic attack of the Lys603 at C2 in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys485A | hydrogen bond donor |
| Glu488A | hydrogen bond acceptor |
| Lys603A | hydrogen bond donor, proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, enzyme-substrate complex formation
Step 8. Lys603 initiates an elimination of water with concomitant deprotonation of itself.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Glu488A | hydrogen bond acceptor |
| Lys603A | covalently attached, hydrogen bond donor |
| His504A(AA) | hydrogen bond acceptor |
| Lys603A | proton donor, electron pair donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, enzyme-substrate complex cleavage, dehydration, intermediate collapse, intermediate formation, schiff base formed
Step 9. Ammonia initiates a nucleophilic attack on the C2 bound to Lys603 in an addition reaction with concomitant deprotonation of the ammonia by Lys603
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Glu488A | hydrogen bond acceptor |
| Lys603A | covalently attached |
| His504A(AA) | hydrogen bond acceptor |
| Lys603A | proton acceptor, electron pair acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation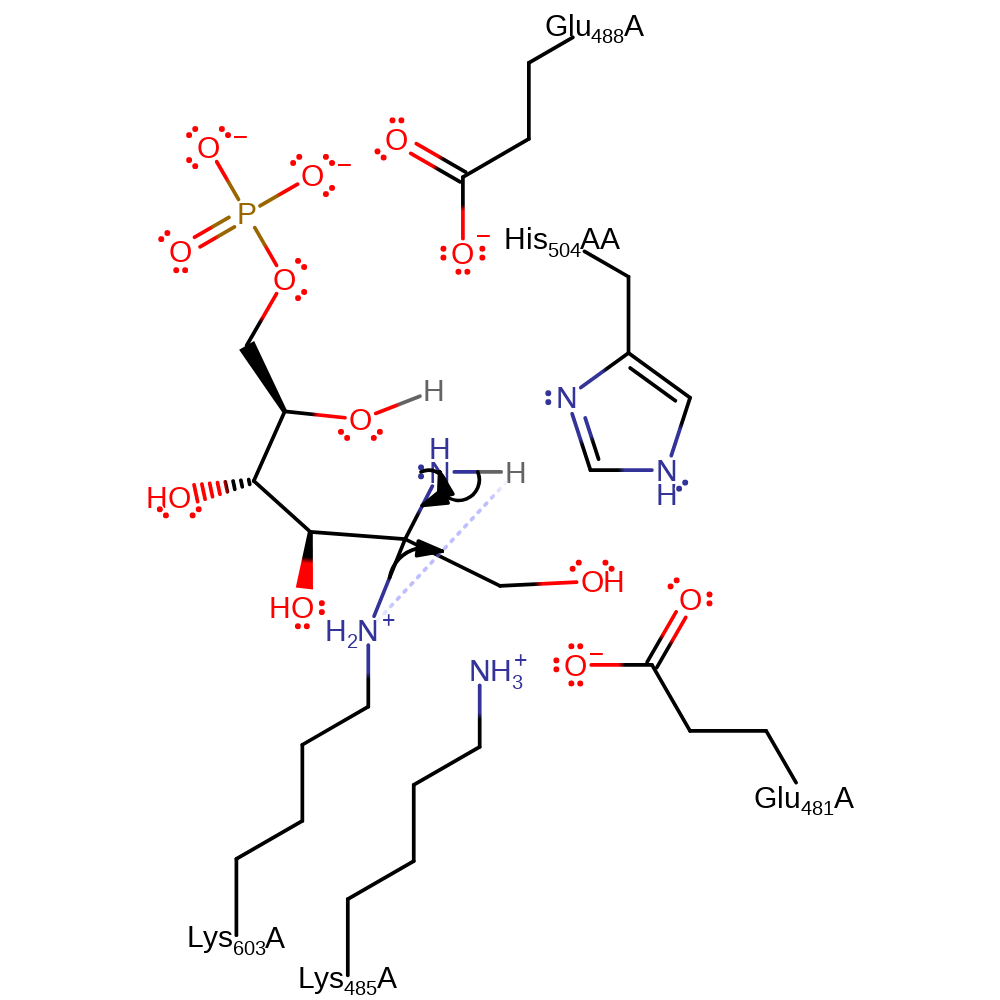
Step 10. The newly bound amine eliminates Lys603 with concomitant deprotonation of itself by Lys603.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Glu488A | hydrogen bond acceptor |
| His504A(AA) | hydrogen bond acceptor |
| Lys603A | proton acceptor, nucleofuge |
Chemical Components
ingold: intramolecular elimination, enzyme-substrate complex cleavage, intermediate formation
Step 11. Glu488 deprotonates the C1 with concomitant double bond rearrangement that deprotonates Lys485.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Glu488A | hydrogen bond acceptor |
| Lys603A | hydrogen bond donor |
| His504A(AA) | hydrogen bond acceptor |
| Lys485A | proton donor |
| Glu488A | proton acceptor |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol), intermediate formation
Step 12. Lys485 deprotonates the C1-OH with concomitant double bond rearrangement and deprotonation of Glu488 by the C2 carbon.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu481A | hydrogen bond acceptor |
| Lys485A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Glu488A | hydrogen bond donor, hydrogen bond acceptor |
| Lys603A | hydrogen bond donor |
| His504A(AA) | hydrogen bond acceptor |
| Lys485A | proton acceptor |
| Glu488A | proton donor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His504A(AA) | hydrogen bond acceptor |
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Glu488A | hydrogen bond acceptor |
| Lys603A | hydrogen bond donor |
| His504A(AA) | proton acceptor |
Chemical Components
proton transfer, intermediate formation
Step 14. The oxyanion initiates a nucleophilic attack on the C1 carbonyl carbon in an addition reaction with concomitant deprotonation of His504AA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His504A(AA) | hydrogen bond donor |
| Lys485A | hydrogen bond donor, electrostatic stabiliser |
| Glu481A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys603A | hydrogen bond donor |
| His504A(AA) | proton donor |






 Download:
Download: