RRNA N-glycosylase
Abrin-A from the seeds of Abrus precatorius is a type II ribosome-inactivating protein (RIP) alongside the homologue Ricin. Like Ricin, it catalyses the endohydrolysis of the N-glycosidic bond at one specific adenosine A4324 on the 28S rRNA. This inhibition of protein biosynthesis is anti-carcinogenic. Abrin-A is composed of two chains which are linked by a disulphide bond. Type I RIPs such as α-Momorcharin however have a single chain, still retaining rRNA-glycosylase activity and share identical catalytic residues to Abrin-A and also excise adenines from a highly conserved loop rRNA.
Reference Protein and Structure
- Sequence
-
P16094
 (3.2.2.22)
(3.2.2.22)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Momordica charantia (balsam apple)

- PDB
-
1ahb
- THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN
(2.2 Å)



- Catalytic CATH Domains
-
4.10.470.10
 3.40.420.10
3.40.420.10  (see all for 1ahb)
(see all for 1ahb)
Enzyme Reaction (EC:3.2.2.22)
Enzyme Mechanism
Introduction
When in the active site, the substrate is in a strained high energy conformation which serves to weaken the C1-N9 bond. N-glycosidic bond cleavage is also facilitated by Arg186 and Ile94, where the resulting ribose oxocarbenium ion is stabilised by Glu183 and nucleophilically attacked by an incoming water molecule which also protonates adenine. The excised adenine and ribose sugar (apart of the ribosome) are free to dissociate from the enzyme, ready for another round of catalysis. Alongside the residues listed here, Tyr70 is important in substrate specificity where it is able to cycle through different conformations throughout the reaction. Experimental evidence suggests this is the most likely mechanism out of the two proposed. The mechanism is based on the protein α-Momorcharin, which has identical catalytic residues to Abrin-A indicating the same mechanism occurs in the two proteins.
Catalytic Residues Roles
| UniProt | PDB* (1ahb) | ||
| Ile94 (main-N) | Ile71A (main-N) | The main chain amide group acts as an electrostatic stabiliser, helping withdraw electrons from the N9-glycosidic bond by forming a hydrogen bond with N1 of adenine. | hydrogen bond donor, electrostatic stabiliser |
| Glu183 | Glu160A | Glu183 stabilises the oxocarbenium ion intermediate. | electrostatic stabiliser |
| Arg186 | Arg163A | Together with Ile94, Arg186 acts as an electrostatic stabiliser by forming a hydrogen bond with N3 of adenine, withdrawing electrons facilitating N9-glycosidic bond cleavage. Arg186 may operate through partial protonation rather than direct due to arginine's high pKa. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
overall reactant used, heterolysis, elimination (not covered by the Ingold mechanisms), proton transfer, bimolecular nucleophilic addition, overall product formedReferences
- Ren J et al. (1994), Structure, 2, 7-16. The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of α-momorcharin. DOI:10.1016/S0969-2126(00)00004-6.
- Tahirov TH et al. (1995), J Mol Biol, 250, 354-367. Crystal Structure of Abrin-a at 2.14 Å. DOI:10.1006/jmbi.1995.0382. PMID:7608980.

Step 1. When in the active site, the substrate is in a strained high energy conformation which serves to weaken the C1-N9 bond. Both Arg186 and Ile94 act as electrostatic stabilisers by hydrogen bonding to N3 and N1 in adenine respectively, withdrawing electrons from this N-glycosidic bond thus facilitating bond cleavage.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu160A | electrostatic stabiliser |
| Arg163A | electrostatic stabiliser |
| Ile71A (main-N) | electrostatic stabiliser |
| Ile71A (main-N) | hydrogen bond donor |
| Arg163A | hydrogen bond donor |
Chemical Components
overall reactant used, heterolysis, elimination (not covered by the Ingold mechanisms)
Step 2. The resulting ribose oxycarbenium ion is stabilised by the positively charged Glu183. Adenine rotates 15° so that the nucleophile, water can enter and attack the oxocarbenium ion. Adenine is protonated by the released proton from water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu160A | electrostatic stabiliser |
| Ile71A (main-N) | hydrogen bond donor |
| Arg163A | hydrogen bond donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall product formedIntroduction
Cleavage of the C1'-N9 bond between adenine and ribose is facilitated by the protonation of N3 by Arg186.The resulting oxycarbonium ion is stabilised by the negatively charged Glu183 residue. The loss of a proton from Arg186 deprotonates and thus activates a water molecule for nucleophilic attack on the C1' atom of the oxycarbonium ion. This is most likely through a concerted mechanism however it has been split into discrete steps to show all correct electron movement. The mechanism seen here uses α-Momorcharin residues but are also conserved in Abrin-A.
Catalytic Residues Roles
| UniProt | PDB* (1ahb) | ||
| Glu183 | Glu160A | Glu183 stabilises the oxocarbenium ion intermediate. | electrostatic stabiliser |
| Arg186 | Arg163A | Arg186 acts as a general acid by protonating the N3 atom of the substrate. It then acts as a general base by deprotonating a water molecule, activating it for nucleophilic attack. | activator, proton acceptor, proton donor |
Chemical Components
proton relay, unimolecular elimination by the conjugate base, proton transfer, overall reactant used, heterolysis, overall product formed, bimolecular nucleophilic addition, native state of enzyme regeneratedReferences
- Monzingo AF et al. (1992), J Mol Biol, 227, 1136-1145. X-ray analysis of substrate analogs in the ricin A-chain active site. DOI:10.1016/0022-2836(92)90526-p. PMID:1433290.
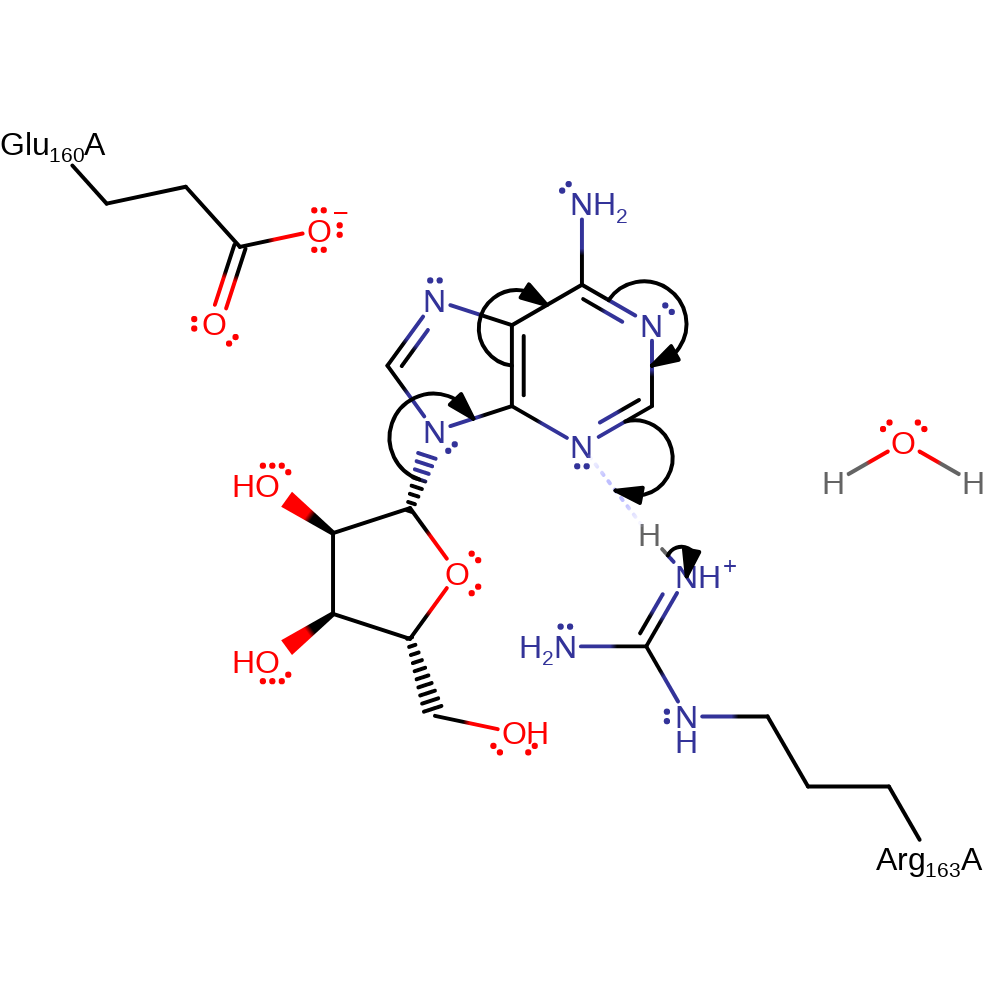
Step 1. The C1'-N9 bond between adenine and ribose is broken, facilitated by the protonation of N3 by Arg186.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu160A | electrostatic stabiliser |
| Arg163A | proton donor |
Chemical Components
proton relay, ingold: unimolecular elimination by the conjugate base, proton transfer, overall reactant used, heterolysis, overall product formed
Step 2. The resulting oxycarbenium ion is stabilised by the negatively charged Glu183 residue. The loss of a proton from Arg186 increases its basicity, causing it to deprotonate a water molecule, activating it for nucleophilic attack on the C1' atom of the oxycarbonium ion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg163A | activator, proton acceptor |
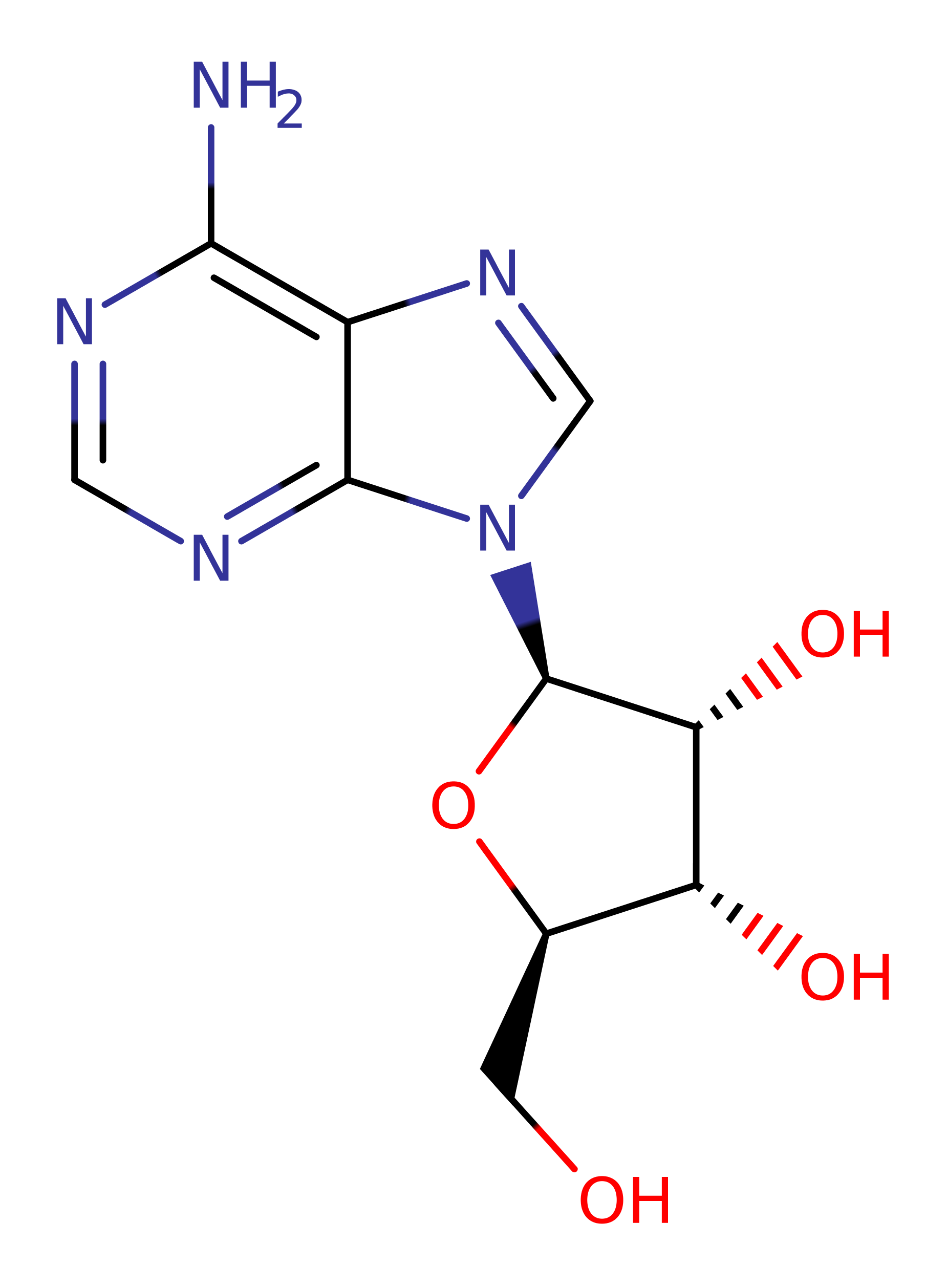

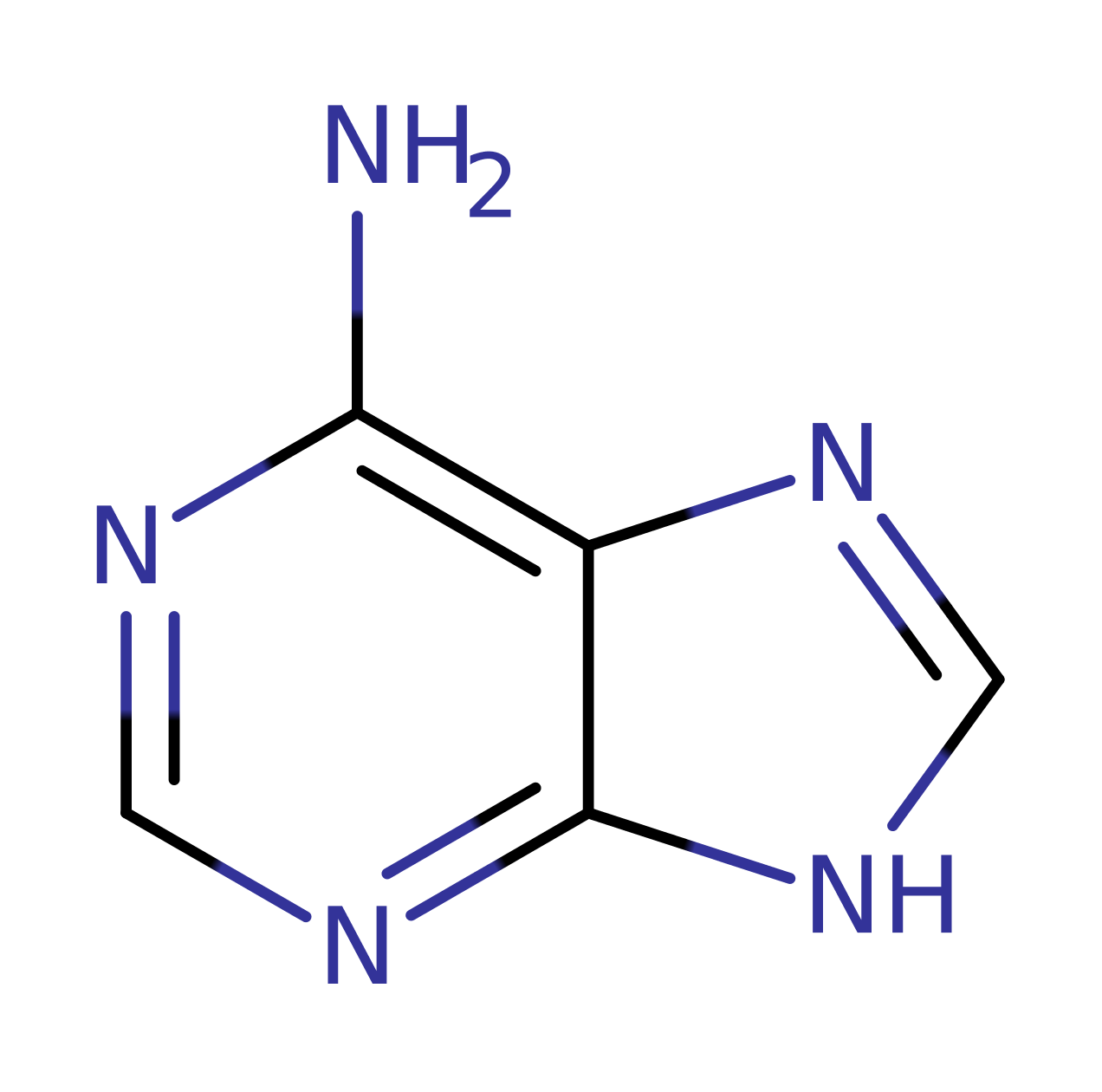

 Download:
Download: 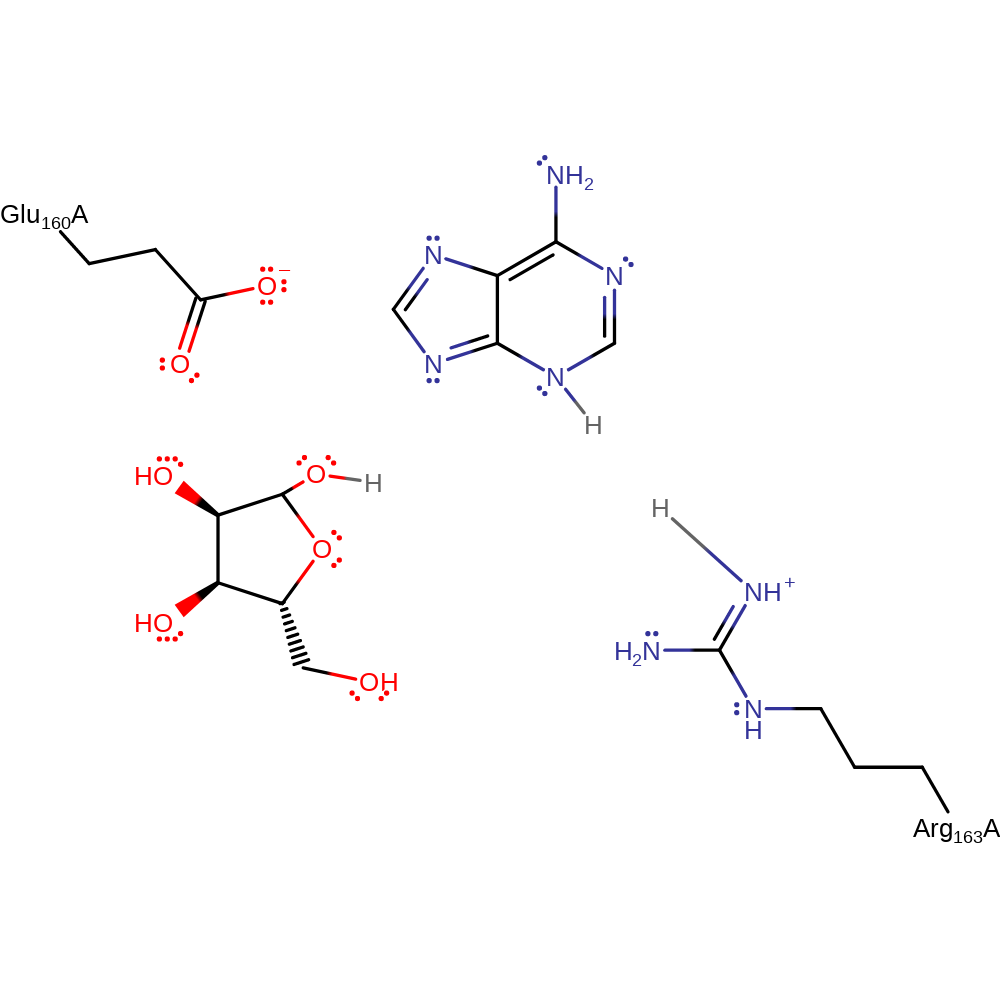 Download:
Download: