Double-stranded uracil-DNA glycosylase
Mismatched base pairing between G and U or G and T is common in DNA replication and would lead to substitution mutations. Therefore the enzyme G/U mismatch-specific DNA glycosylase is vital in preserving the integrity of DNA: it is able to hydrolyse the glycosidic bond to release the uracil or thymine base, thus creating an abasic site which allows the entire nucleotide to be recognised and removed. The enzyme shows significant structural and functional similarity to the well characterised family of Uracil DNA glycosylases which remove uracil wherever it occurs in DNA; however there is very little sequence similarity suggesting that convergent evolution may be responsible for the similarities observed. It is apart of the MUG/TDGs family of uracil-DNA glycosylases along with homologue human thymine-DNA glycosylase of which, has two alternative mechanisms proposed.
Reference Protein and Structure
- Sequence
-
Q13569
 (3.2.2.29)
(3.2.2.29)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
3ufj
- Human Thymine DNA Glycosylase Bound to Substrate Analog 2'-fluoro-2'-deoxyuridine
(2.967 Å)



- Catalytic CATH Domains
-
3.40.470.10
 (see all for 3ufj)
(see all for 3ufj)
- Cofactors
- Water (1)
Enzyme Reaction (EC:3.2.2.29)
Enzyme Mechanism
Introduction
The below proposed mechanism is termed the 'Histidine Mechanism'. It is favoured of the two due to the lower calculated free energy of the rate determining step (N-glycosidic bond cleavage) than the alternative, 'direct mechanism'. That being said, there is also a suggestion the actual mechanism is a hybrid of the two. His151 protonates the uracil, facilitating the cleavage of the N-glycosidic bond. Subsequently, water held in position by Asp140 nucleophilically attacks the ribose intermediate. Further proton transfer from this water and then back to His151 from excised uracil sees the termination of the reaction mechanism. Enzyme reaction rates observed experimentally are low for this enzyme, which are due to lower levels of protonated His151, needed to lower the free energy barrier relative to unprotonated His151 seen in most enzyme complexes.
Catalytic Residues Roles
| UniProt | PDB* (3ufj) | ||
| Asn140 | Asn140(36)A | Asn140's role is to hydrogen bond thus position the water for nucleophilic attack. | electrostatic interaction |
| His151 | His151(47)A | His151 reduces the free energy barrier for N-glycosydic cleavage in being firstly protonated and subsequently protonating the thymine base (reprotonated at the end of the reaction). The proton helps stabilise the increasingly negative charge on the N transition state during N-glycosidic bond cleavage. | proton acceptor, proton donor |
Chemical Components
proton transfer, proton relay, overall reactant used, intermediate formation, heterolysis, unimolecular elimination by the conjugate base, rate-determining step, intermediate collapse, overall product formed, charge delocalisation, bimolecular nucleophilic addition, intermediate terminated, native state of enzyme regeneratedReferences
- Kanaan N et al. (2015), J Phys Chem B, 119, 12365-12380. Mechanism of the Glycosidic Bond Cleavage of Mismatched Thymine in Human Thymine DNA Glycosylase Revealed by Classical Molecular Dynamics and Quantum Mechanical/Molecular Mechanical Calculations. DOI:10.1021/acs.jpcb.5b05496. PMID:26320595.
- Hardeland U et al. (2000), J Biol Chem, 275, 33449-33456. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. DOI:10.1074/jbc.M005095200. PMID:10938281.
- Barrett TE et al. (1998), Cell, 92, 117-129. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. PMID:9489705.
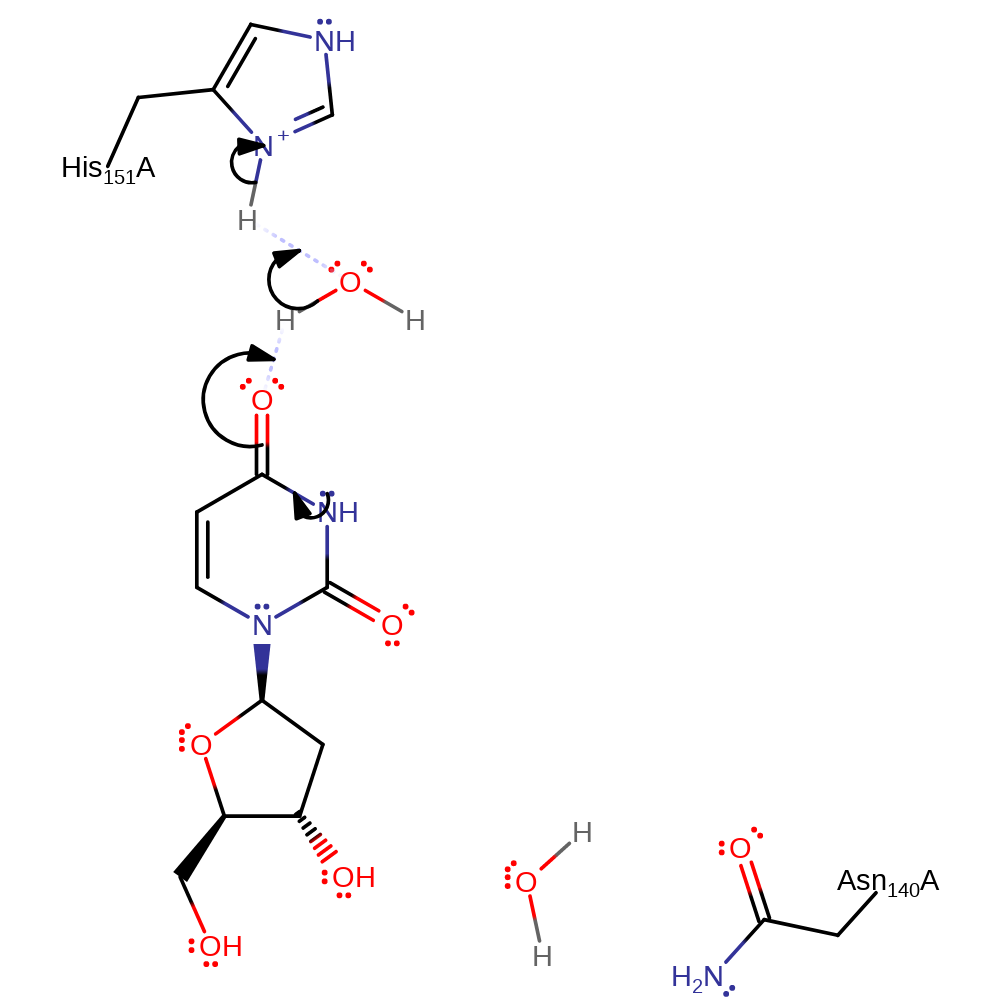
Step 1. Proton transfer from protonated His151 to a carbonyl on the uracil via a water molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His151(47)A | proton donor |
Chemical Components
proton transfer, proton relay, overall reactant used, intermediate formation
Step 2. N-glycosidic bond cleavage releases the recently protonated uracil. This results in the formation of an oxacarbenium ion and anionic base.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
heterolysis, ingold: unimolecular elimination by the conjugate base, rate-determining step, intermediate collapse, overall product formed, charge delocalisation
Step 3. Nucleophilic attack by water on the oxacarbenium ion. There is also subsequent proton transfer from the same water to uracil after rearrrangement of thymine relative to the sugar.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn140(36)A | electrostatic interaction |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate collapse, overall product formed
Step 4. Back transfer of proton from thymine to His151 via a water molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His151(47)A | proton acceptor |
Chemical Components
proton relay, proton transfer, overall product formed, intermediate terminated, native state of enzyme regenerated, charge delocalisationIntroduction
The below mechanism is termed the 'direct mechanism'. First, N-glycosidic bond cleavage in the presence of protonated His-151 results in an oxacarbenium ion that is then nucleophilically attacked by a water molecule (which is held in position by Asn140 via hydrogen bonds). Proton transfer from the hydronium ion to the excised uracil marks the end of the reaction and the enzyme is ready for another excision reaction.
Catalytic Residues Roles
| UniProt | PDB* (3ufj) | ||
| Asn140 | Asn140(36)A | Asn140's role is to hydrogen bond thus position the water for nucleophilic attack. | electrostatic interaction |
| His151 | His151(47)A | Protonated His151 enables the rate determining step to occur at a lower energy. The proton helps stabilise the increasingly negative charge on the N transition state during N-glycosidic bond cleavage. |
Chemical Components
rate-determining step, heterolysis, elimination (not covered by the Ingold mechanisms), overall reactant used, intermediate formation, bimolecular nucleophilic addition, overall product formed, intermediate terminated, proton transferReferences
- Kanaan N et al. (2015), J Phys Chem B, 119, 12365-12380. Mechanism of the Glycosidic Bond Cleavage of Mismatched Thymine in Human Thymine DNA Glycosylase Revealed by Classical Molecular Dynamics and Quantum Mechanical/Molecular Mechanical Calculations. DOI:10.1021/acs.jpcb.5b05496. PMID:26320595.
- Hardeland U et al. (2000), J Biol Chem, 275, 33449-33456. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. DOI:10.1074/jbc.M005095200. PMID:10938281.
- Barrett TE et al. (1998), Cell, 92, 117-129. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. PMID:9489705.

Step 1. Cleavage of the N-glycosidic bond to form an oxacarbenium ion and negatively charged base.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn140(36)A | electrostatic interaction |
Chemical Components
rate-determining step, heterolysis, elimination (not covered by the Ingold mechanisms), overall reactant used, intermediate formation
Step 2. Nucleophilic attack of the oxacarbenium ion by a water molecule. The sugar intermediate reorientates in relation to the excised uracil. This brings the nitrogen on uracil closer to the proton on the hydronium ion for proton transfer to the uracil.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn140(36)A | electrostatic interaction |




 Download:
Download:  Download:
Download: