Phosphoenolpyruvate carboxylase
Phosphoenolpyruvate carboxylase (PEPC) catalyses the irreversible carboxylation of phosphoenolpyruvate (PEP) to form oxaloacetate (OAA) and Pi using divalent Mg(II) or Mn(II) as a cofactor. The active species for initial carboxylation is not carbon dioxide but the chemically less reactive bicarbonate anion, which dominates in the cytoplasm where PEPC functions. The enzyme is present in all photosynthetic organisms, including higher plants, green algae, cyanobacteria and photosynthetic bacteria, and most nonphotosynthetic bacteria and protozoa, but is notably absent from animals, fungi, and yeasts. PEPC primarily plays an anaplerotic role by replenishing C4-dicarboxylic acids utilised for both energy and biosynthetic metabolisms.
Reference Protein and Structure
- Sequence
-
P00864
 (4.1.1.31)
(4.1.1.31)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1qb4
- CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE
(2.6 Å)



- Catalytic CATH Domains
- (see all for 1qb4)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:4.1.1.31)
Enzyme Mechanism
Introduction
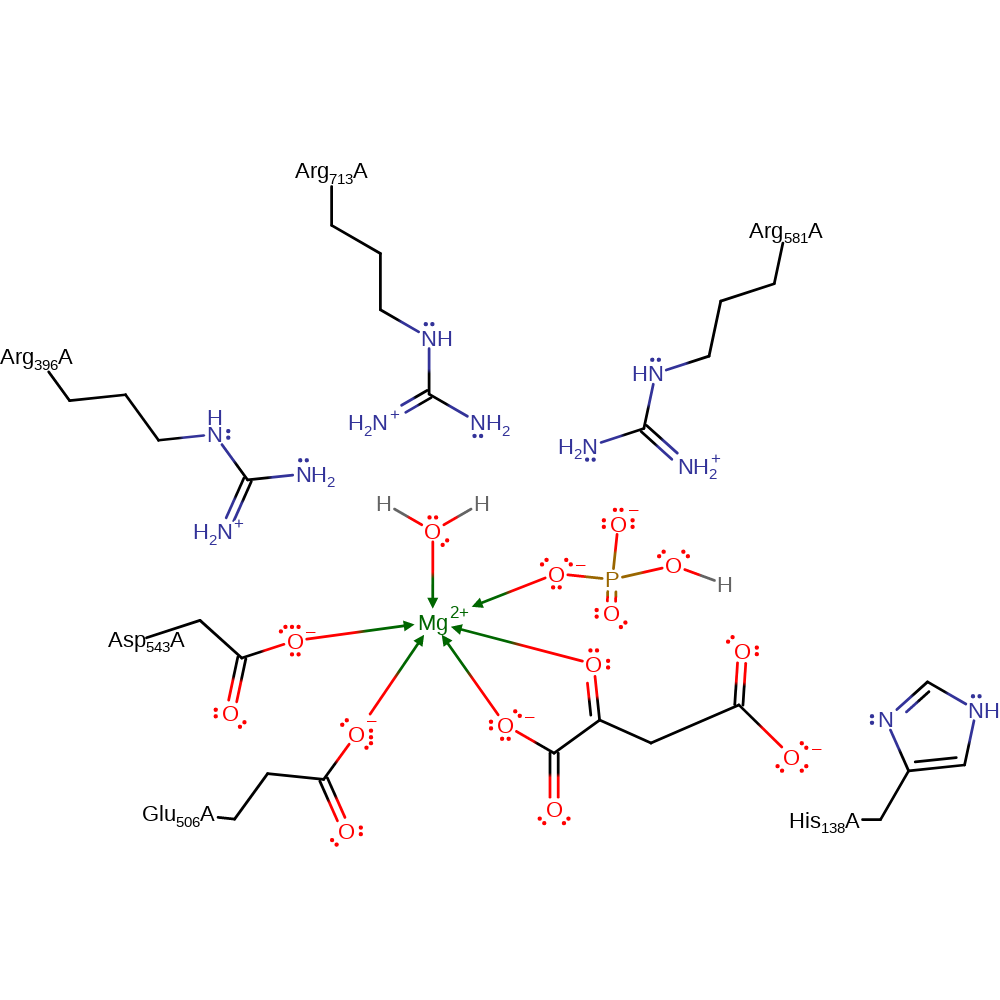
The first chemical step in the proposed mechanism is the nucleophilic attack by bicarbonate to form carboxyphosphate and the enolate of pyruvate. In order to make the phosphorus atom less negative, the positively charged electrostatic pocket formed by three arginine residues allows the dissipation of the negative charges of the phosphate group during the approach. In the second chemical step of the proposed mechanism His138 acts as a base to abstract a proton from carboxyphosphate; CO2 is generated, the hydrophobic environment in the active site pocket may stabilise the CO2 liberated from the carboxyphosphate. Finally the carbon dioxide generated is nucleophilically attacked by enolate of pyruvate to generate oxaloacetate. The phosphate anion abstracts a proton from His138 to form a phosphate group.
Catalytic Residues Roles
| UniProt | PDB* (1qb4) | ||
| Glu506, Asp543 | Glu506A, Asp543A | Coordinate the stabilizing metal | metal ligand |
| His138 | His138A | Acts as a general acid/base. | proton acceptor, proton donor |
| Arg581, Arg713, Arg396 | Arg581A, Arg713A, Arg396A | The positively charged electrostatic pocket formed by Arg396, Arg699, and Arg713 allows the dissipation of the negative charges of the phosphate group during the approach | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic addition, proton transfer, overall reactant used, bimolecular nucleophilic substitution, decarboxylation, aldol addition, overall product formed, native state of enzyme regeneratedReferences
- Matsumura H et al. (2002), Structure, 10, 1721-1730. Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. PMID:12467579.
- Kai Y et al. (2003), Arch Biochem Biophys, 414, 170-179. Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. PMID:12781768.
- Terada K et al. (1991), Eur J Biochem, 202, 797-803. Site-directed mutagenesis of the conserved histidine residue of phosphoenolpyruvate carboxylase. His138 is essential for the second partial reaction. PMID:1765093.
Step 1. There is nucleophilic attack from the water molecule to the CO2 molecule forming carbonicacid
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg396A | electrostatic stabiliser |
| Arg581A | electrostatic stabiliser |
| Arg713A | electrostatic stabiliser |
| Glu506A | metal ligand |
| Asp543A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic additionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg396A | electrostatic stabiliser |
| Arg581A | electrostatic stabiliser |
| Arg713A | electrostatic stabiliser |
| Glu506A | metal ligand |
| Asp543A | metal ligand |
Chemical Components
proton transfer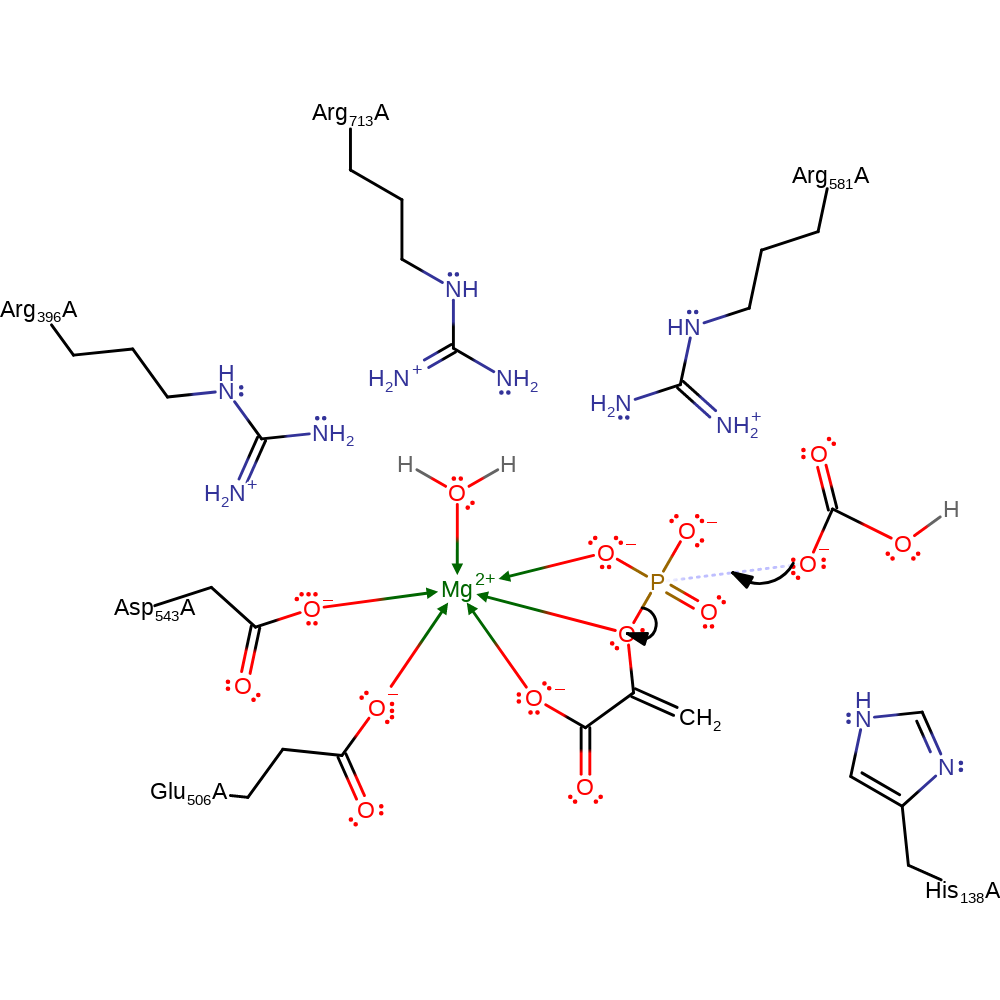
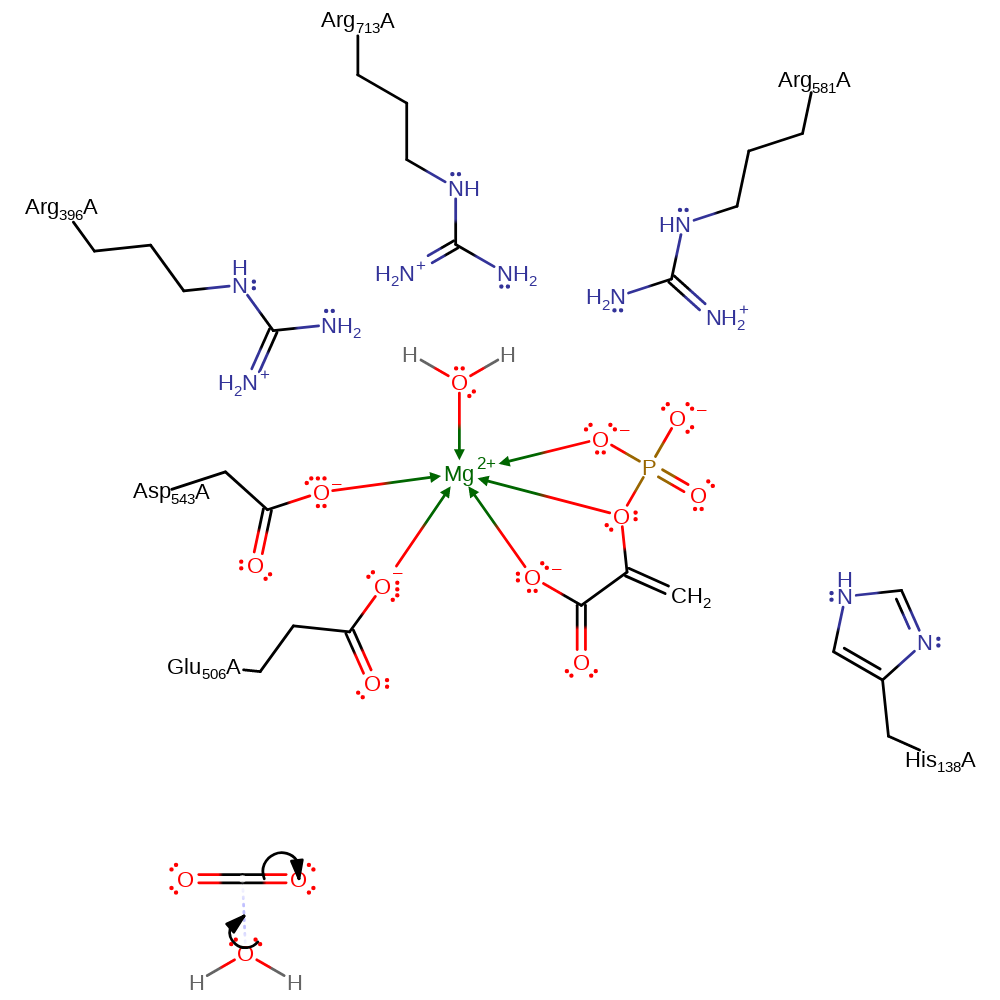
Step 3. Nucleophilic attack by bicarbonate forms carboxyphosphate and the enolate of pyruvate. In order to make the phosphorus atom less negative, the positively charged electrostatic pocket formed by the arginine residues allows the dissipation of the negative charges of the phosphate group during the approach
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg396A | electrostatic stabiliser |
| Arg581A | electrostatic stabiliser |
| Arg713A | electrostatic stabiliser |
| Glu506A | metal ligand |
| Asp543A | metal ligand |
Chemical Components
overall reactant used, ingold: bimolecular nucleophilic substitution
Step 4. His 138 acts as a general base abstracting a proton from the carboxyphosphate, this leads to CO2 being released.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg396A | electrostatic stabiliser |
| Arg581A | electrostatic stabiliser |
| Arg713A | electrostatic stabiliser |
| Glu506A | metal ligand |
| Asp543A | metal ligand |
| His138A | proton acceptor |
Chemical Components
decarboxylation, proton transfer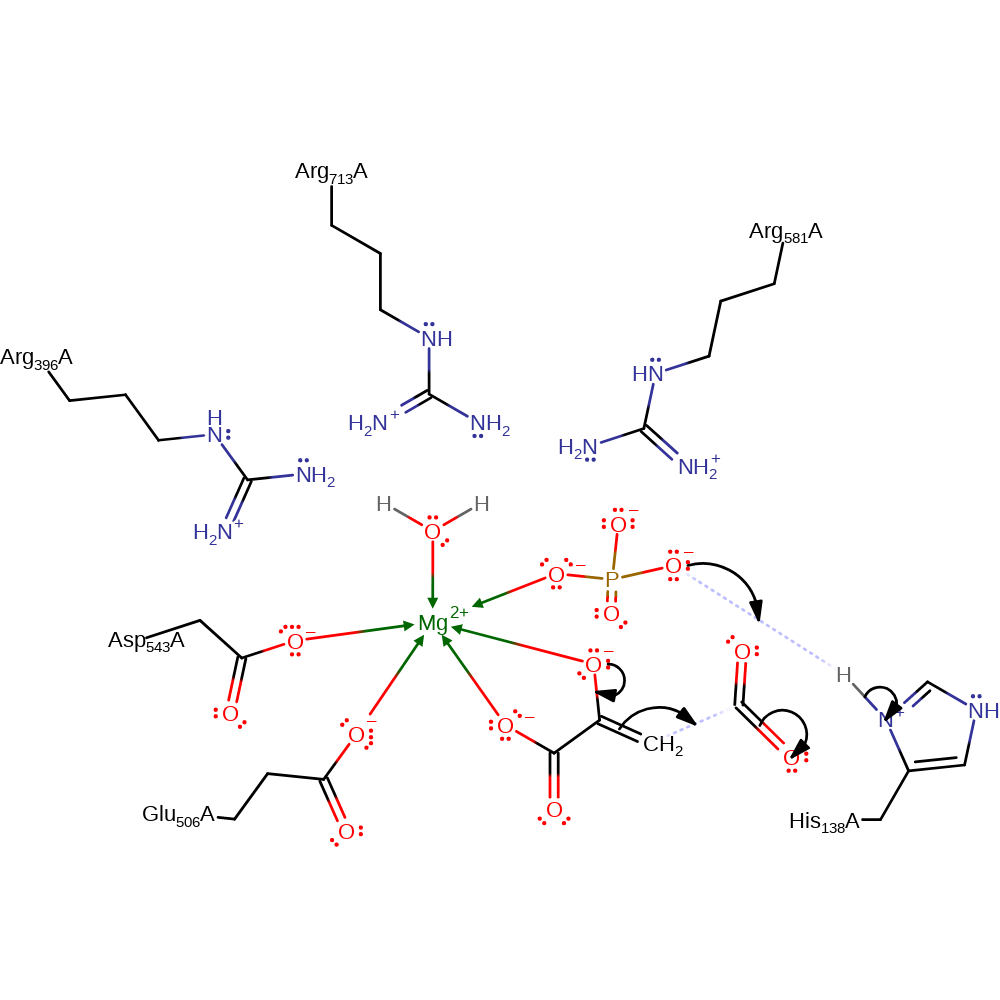
Step 5. An aldol addition leads to the phosphoenolpyruvate being carboxylated. The phosphate accepts a proton from His 138.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg396A | electrostatic stabiliser |
| Arg581A | electrostatic stabiliser |
| Arg713A | electrostatic stabiliser |
| Glu506A | metal ligand |
| Asp543A | metal ligand |
| His138A | proton donor |






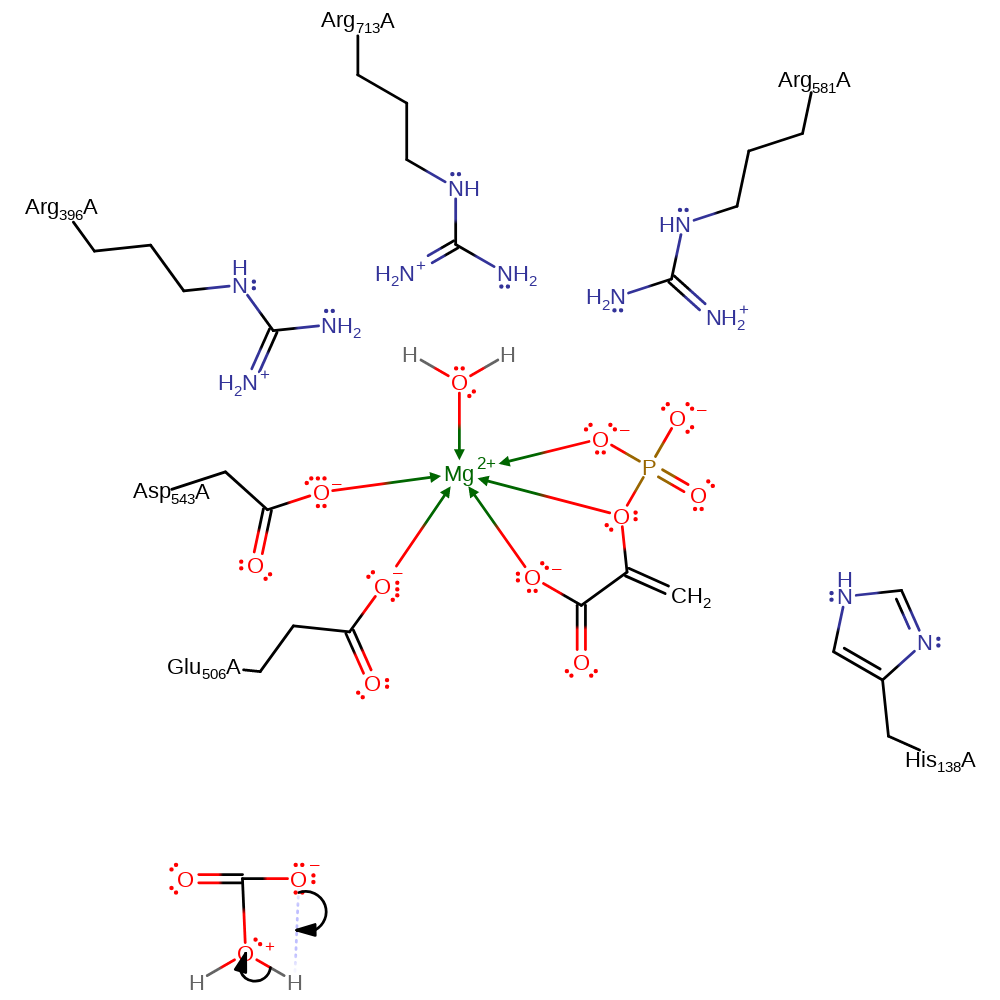
 Download:
Download: