UDP-glucose-hexose-1-phosphate uridylyltransferase
Nucleotidyltransferases catalyse the covalent modification of a variety of biological molecules. These reactions are crucial for the synthesis of coenzymes, cyclic nucleotides, polynucleotides, and nucleotide sugars. These reactions involve substitutions at the R -phosphorus of a nucleotidyl donor substrate and result in displacement of a phosphoryl ester or pyrophosphate. Substrates for such reactions may include nucleoside di- or triphosphates, as well as nucleotide sugars, such as UDP-Glc. Galactose-1-phosphate uridylyltransferase (hexose-1-phosphate uridylyltransferase) catalyses the exchange of the UMP moiety between the hexose 1-phosphates of Glc and Gal and their corresponding UDP-sugar. The enzyme is distinct among nucleotidyl transferases that use phosphates as acceptor groups in that it is the only one that does not utilise nucleoside di- or triphosphates as the nucleotidyl donor substrate. The reaction is part of the Leloir pathway of galactose metabolism required for the normal equilibration of UDP-hexoses among most organisms. Deficiencies in uridylyltransferase activity culminate in the metabolic disease galactosemia, which occurs as an autosomal recessive trait.
Reference Protein and Structure
- Sequence
-
P09148
 (2.7.7.12)
(2.7.7.12)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1hxq
- THE STRUCTURE OF NUCLEOTIDYLATED GALACTOSE-1-PHOSPHATE URIDYLYLTRANSFERASE FROM ESCHERICHIA COLI AT 1.86 ANGSTROMS RESOLUTION
(1.86 Å)



- Catalytic CATH Domains
-
3.30.428.10
 (see all for 1hxq)
(see all for 1hxq)
- Cofactors
- Zinc(2+) (1) Metal MACiE
Enzyme Reaction (EC:2.7.7.12)
Enzyme Mechanism
Introduction
His166 in the active site of the the enzyme in Escherichia coli attacks the alpha-phosphorus of UDP--Glc, displaces Glc-1-P, and forms the high energy covalent uridylyl-enzyme (UMP-enzyme) intermediate, which then reacts with Gal-1-P to produce UDP-Gal.
Catalytic Residues Roles
| UniProt | PDB* (1hxq) | ||
| His164 (main-C) | His164A (main-C) | Along with it's side chain, this residue is important for positioning and stabilising the catalytic His166. | activator |
| Ser161 | Ser161A | Activates and stabilises the reaction intermediates. | hydrogen bond donor, electrostatic stabiliser, steric role |
| His166 | His166A | Acts as the catalytic nucleophile. | hydrogen bond donor, nucleophile, nucleofuge |
| His164 | His164A | Forms part of the zinc binding site. Also involved in maintaining the correct positioning of the substrates in the active site. | hydrogen bond acceptor, metal ligand, steric role |
| Cys52, Cys55, His115 | Cys52A, Cys55A, His115A | Forms part of the zinc binding site. | metal ligand |
| Asn153, Gln168, His166 (main-C) | Asn153A, Gln168A, His166A (main-C) | Holds the catalytically important water in position. These interactions help stabilise the phosphate intermediate. | activator, hydrogen bond donor |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, overall product formed, intermediate formation, enzyme-substrate complex cleavage, intermediate terminated, native state of enzyme regeneratedReferences
- Wedekind JE et al. (1996), Biochemistry, 35, 11560-11569. The Structure of Nucleotidylated Histidine-166 of Galactose-1-phosphate Uridylyltransferase Provides Insight into Phosphoryl Group Transfer†,‡. DOI:10.1021/bi9612677. PMID:8794735.
- McCorvie TJ et al. (2011), IUBMB Life, 63, 694-700. The structural and molecular biology of type I galactosemia: Enzymology of galactose 1-phosphate uridylyltransferase. DOI:10.1002/iub.511. PMID:21793161.
- Geeganage S et al. (2002), Methods Enzymol, 134-148. Galactose-1-Phosphate Uridylyltransferase: Kinetics of Formation and Reaction of Uridylyl-Enzyme Intermediate in Wild-Type and Specifically Mutated Uridylyltransferases. DOI:10.1016/s0076-6879(02)54010-6.
- Brenner C (2002), Biochemistry, 41, 9003-9014. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. PMID:12119013.
- Geeganage S et al. (2000), Biochemistry, 39, 5397-5404. Roles of Two Conserved Amino Acid Residues in the Active Site of Galactose-1-Phosphate Uridylyltransferase: An Essential Serine and a Nonessential Cysteine†. DOI:10.1021/bi992594s. PMID:10820011.
- Geeganage S et al. (1999), Biochemistry, 38, 13398-13406. Significance of Metal Ions in Galactose-1-Phosphate Uridylyltransferase: An Essential Structural Zinc and a Nonessential Structural Iron†. DOI:10.1021/bi9910631. PMID:10529216.
- Thoden JB et al. (1997), Biochemistry, 36, 1212-1222. Structural Analysis of the H166G Site-Directed Mutant of Galactose-1-phosphate Uridylyltransferase Complexed with either UDP-glucose or UDP-galactose: Detailed Description of the Nucleotide Sugar Binding Site†,‡. DOI:10.1021/bi9626517. PMID:9063869.
- Wedekind JE et al. (1995), Biochemistry, 34, 11049-11061. Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 A resolution. PMID:7669762.
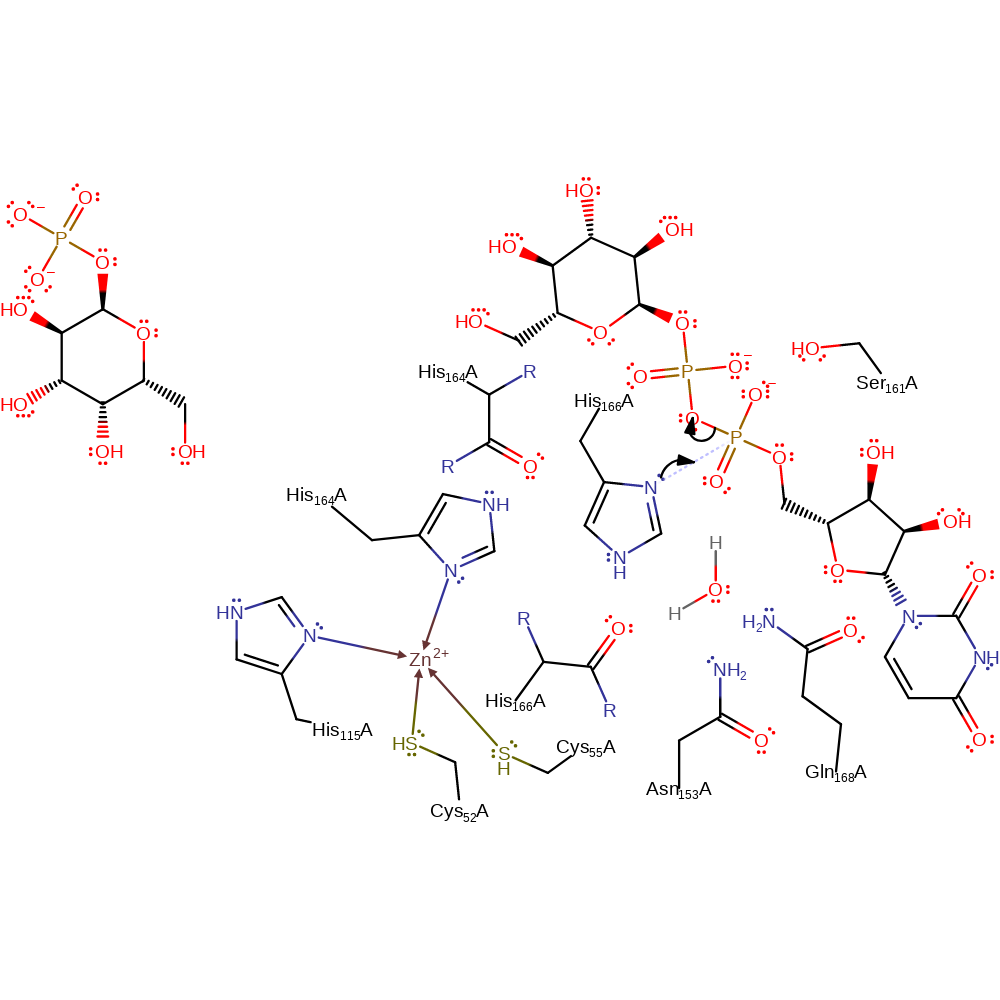
Step 1. His166 initiates a nucleophilic attack on the alpha-phosphate of the UDP-glucose substrate in a substitution reaction, eliminating glucose phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn153A | hydrogen bond donor, activator |
| Ser161A | hydrogen bond donor, electrostatic stabiliser, steric role |
| His164A | metal ligand, steric role, hydrogen bond acceptor |
| His166A | hydrogen bond donor |
| Gln168A | electrostatic stabiliser, hydrogen bond donor |
| Cys52A | metal ligand |
| Cys55A | metal ligand |
| His115A | metal ligand |
| His164A (main-C) | activator |
| His166A (main-C) | activator |
| His166A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, overall product formed, intermediate formation
Step 2. Galactose phosphate initiates a nucleophilic attack on the alpha-phosphate of the covalently bound UDP in a substitution reaction, eliminating His166.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn153A | hydrogen bond donor, activator |
| Ser161A | hydrogen bond donor, electrostatic stabiliser, steric role |
| His164A | metal ligand, steric role, hydrogen bond acceptor |
| His166A | hydrogen bond donor |
| Gln168A | electrostatic stabiliser, hydrogen bond donor |
| Cys52A | metal ligand |
| Cys55A | metal ligand |
| His115A | metal ligand |
| His166A | nucleofuge |



 Download:
Download: