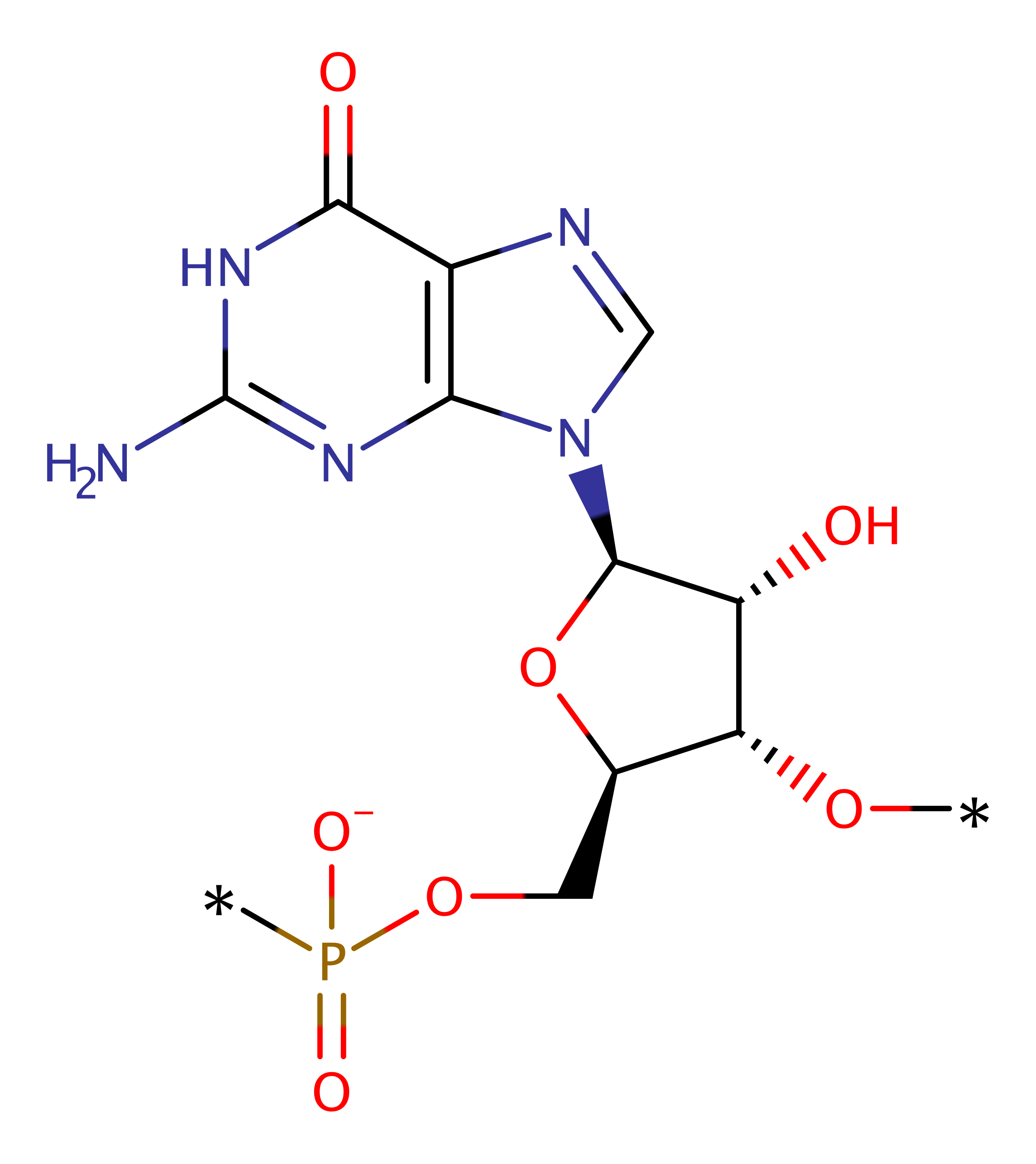
TRNA-guanine transglycosylase
A variety of modified bases are found in RNAs and around 1 % of the Escherichia coli genome codes for proteins involved in tRNA modification. The hypermodified base quenine, a 7-deazaguanine derivative, replaces guanine-34 at the wobble position of tRNAs specific for Asn, Asp, His and Tyr. In vitro and in vivo studies showed that quenine in tRNAs affects codon selection and prevents stop codon readthrough of tobacco mosaic virus RNA in a codon context-dependent manner. However, its general function remains not very well understood [PMID:8654383]. Quenine is synthesised de novo in prokaryotes, whereas it is a nutrient for eukaryotes. In prokaryotes, the quenine biosynthesis requires several steps, not all of them yet known [PMID:8654383]. The biosynthesis begins outside of the tRNA with the conversion of GTP to the quenine precursor 7-aminomethyl-7-deazaguanine (preQ1). tRNA-guanine transglycosylase (TGT) replaces the guanine-34 at the wobble position of the cognate tRNAs with preQ1. tRNA-preQ1 is modified further by the S-adenosylmethionine-requiring enzyme QueA and an unknown vitamin B12-dependent enzyme leading to quenine [PMID:8654383].
Reference Protein and Structure
- Sequence
-
P28720
 (2.4.2.29)
(2.4.2.29)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Zymomonas mobilis subsp. mobilis ZM4 = ATCC 31821 (Bacteria)

- PDB
-
1pud
- TRNA-GUANINE TRANSGLYCOSYLASE
(1.85 Å)



- Catalytic CATH Domains
-
3.20.20.105
 (see all for 1pud)
(see all for 1pud)
- Cofactors
- Zinc(2+) (1)
Enzyme Reaction (EC:2.4.2.29)
Enzyme Mechanism
Introduction
The catalytic mechanism proposed on the basis of the crystal structure determination of TGT: (1) The carboxylate group of Asp 280 first attacks the wobble guanosine at the C1' atom, leading to guanine release and formation of a covalent bond between the aspartate side chain and the ribose in an alpha-configuration. (2) The second reaction catalysed by TGT is the substitution of the aspartate 280 by preQ1, leading to the formation of the final preQ1-modified tRNA. It has been proposed that deprotonation of preQ1 nitrogen N9 occurs before it attacks the C1' atom.
Catalytic Residues Roles
| UniProt | PDB* (1pud) | ||
| Asp280 | Asp280A | Acts as a nucleophile. | covalent catalysis |
| Cys318, Cys320, Cys323, His349 | Cys318A, Cys320A, Cys323A, His349A | Forms part of the zinc binding site. | metal ligand |
| Asp102 | Asp102A | Acts as a general acid/base. | proton shuttle (general acid/base) |
Chemical Components
References
- Xie W et al. (2003), Nat Struct Biol, 10, 781-788. Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate. DOI:10.1038/nsb976. PMID:12949492.
- Barandun LJ et al. (2015), Chemistry, 21, 126-135. Replacement of water molecules in a phosphate binding site by furanoside-appended lin-benzoguanine ligands of tRNA-guanine transglycosylase (TGT). DOI:10.1002/chem.201405764. PMID:25483606.
- Biela I et al. (2013), PLoS One, 8, e64240-. Investigation of specificity determinants in bacterial tRNA-guanine transglycosylase reveals queuine, the substrate of its eucaryotic counterpart, as inhibitor. DOI:10.1371/journal.pone.0064240. PMID:23704982.
- Stengl B et al. (2005), Chembiochem, 6, 1926-1939. Mechanism and substrate specificity of tRNA-guanine transglycosylases (TGTs): tRNA-modifying enzymes from the three different kingdoms of life share a common catalytic mechanism. DOI:10.1002/cbic.200500063. PMID:16206323.
- Brenk R et al. (2003), Chembiochem, 4, 1066-1077. Flexible adaptations in the structure of the tRNA-modifying enzyme tRNA-guanine transglycosylase and their implications for substrate selectivity, reaction mechanism and structure-based drug design. DOI:10.1002/cbic.200300644. PMID:14523925.
- Grädler U et al. (1999), FEBS Lett, 454, 142-146. Mutagenesis and crystallographic studies ofZymomonas mobilistRNA-guanine transglycosylase to elucidate the role of serine 103 for enzymatic activity. DOI:10.1016/s0014-5793(99)00793-0. PMID:10413112.
- Romier C et al. (1996), Biochemistry, 35, 15734-15739. Mutagenesis and Crystallographic Studies ofZymomonas mobilistRNA-Guanine Transglycosylase Reveal Aspartate 102 as the Active Site Nucleophile†,‡. DOI:10.1021/bi962003n. PMID:8961936.
- Romier C et al. (1996), EMBO J, 15, 2850-2857. Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange. PMID:8654383.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp102A | proton shuttle (general acid/base) |
| Asp280A | covalent catalysis |
| Cys318A | metal ligand |
| Cys320A | metal ligand |
| Cys323A | metal ligand |
| His349A | metal ligand |