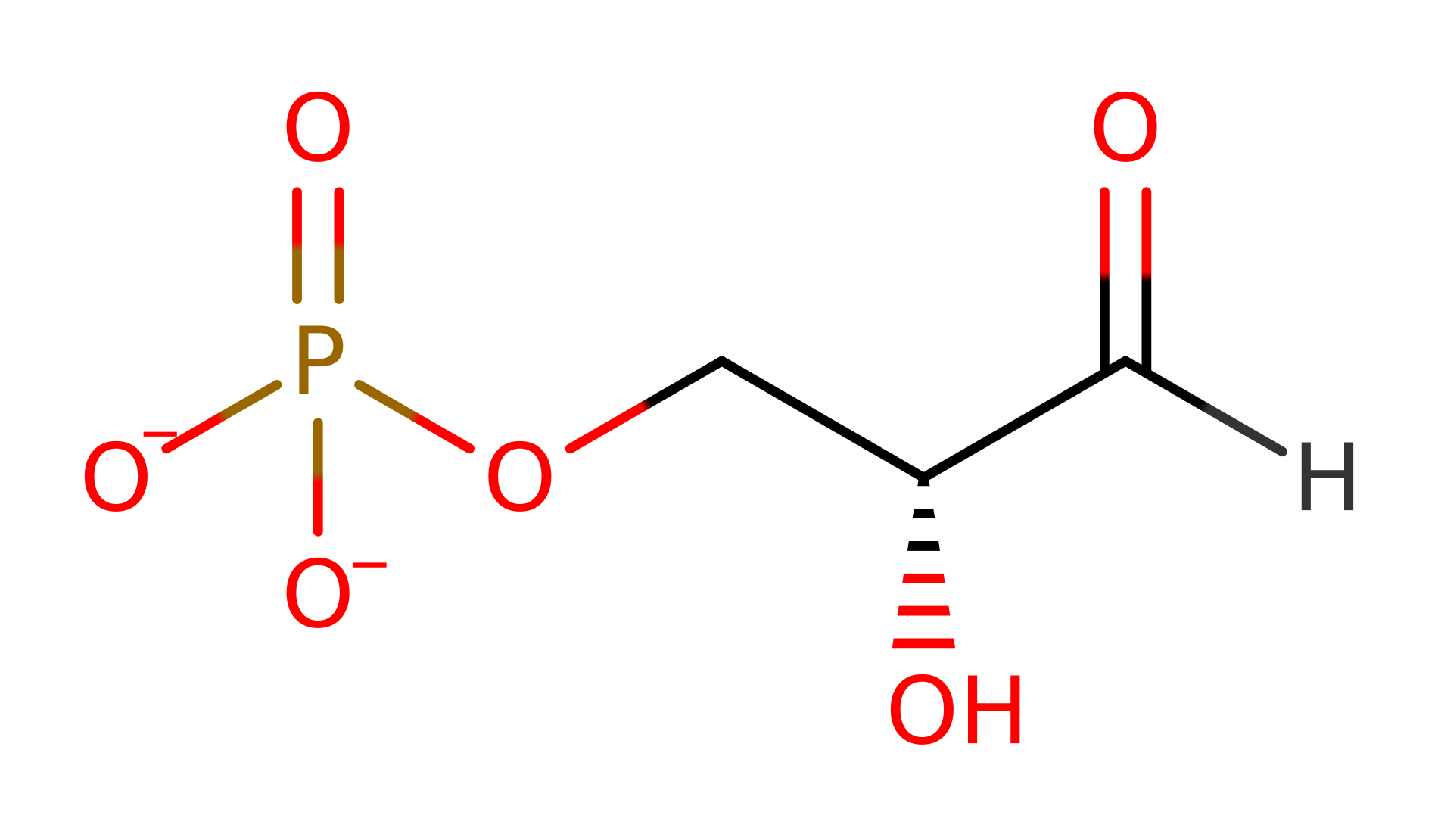
Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) (type I)
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is the enzyme responsible for the interconversion of 1,3-diphosphoglycerate and glyceraldehyde-3-phosphate, a central step in glycolysis and gluconeogenesis.
Reference Protein and Structure
- Sequence
-
P56649
 (1.2.1.12)
(1.2.1.12)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Panulirus versicolor (painted spiny lobster)

- PDB
-
1szj
- STRUCTURE OF HOLO-GLYCERALDEHYDE-3-PHOSPHATE-DEHYDROGENASE FROM PALINURUS VERSICOLOR REFINED 2.0 ANGSTROM RESOLUTION
(2.0 Å)



- Catalytic CATH Domains
-
3.30.360.10
 (see all for 1szj)
(see all for 1szj)
Enzyme Reaction (EC:1.2.1.12)
+
+
→
+
+
Alternative enzyme names: 3-phosphoglyceraldehyde dehydrogenase, NAD-dependent glyceraldehyde phosphate dehydrogenase, NADH-glyceraldehyde phosphate dehydrogenase, Dehydrogenase, glyceraldehyde phosphate, Glyceraldehyde phosphate dehydrogenase (NAD), Glyceraldehyde-3-P-dehydrogenase, Glyceraldehyde-3-phosphate dehydrogenase (NAD), Phosphoglyceraldehyde dehydrogenase, Triosephosphate dehydrogenase, NAD-dependent glyceraldehyde-3-phosphate dehydrogenase, GAPDH,
Enzyme Mechanism
Introduction
The mechanism for this enzyme is relatively poorly understood. It is thought that the reaction proceeds through a covalent intermediate between the substrate and Cys149. The final step of the reaction is a phosphorylation, after which the product dissociates from the active site.
Catalytic Residues Roles
| UniProt | PDB* (1szj) | ||
| Cys148 | Cys149(148)G(A) | Acts as the catalytic nucleophile. | covalent catalysis, proton shuttle (general acid/base) |
| His175 | His176(175)G(A) | Acts as a general acid/base to activate the thiol group during catalysis. | proton shuttle (general acid/base) |
*PDB label guide - RESx(y)B(C) - RES: Residue Name; x: Residue ID in PDB file;
y: Residue ID in PDB sequence if different from PDB file; B: PDB Chain;
C: Biological Assembly Chain if different from PDB. If label is "Not Found" it means this residue is not found in the reference PDB.
Chemical Components
References
- Song SY et al. (1999), J Mol Biol, 287, 719-725. Structure of active site carboxymethylated D-glyceraldehyde-3-phosphate dehydrogenase from Palinurus versicolor. DOI:10.1006/jmbi.1999.2628. PMID:10191140.
- Shen YQ et al. (2002), Acta Crystallogr D Biol Crystallogr, 58, 1287-1297. Structures of D-glyceraldehyde-3-phosphate dehydrogenase complexed with coenzyme analogues. PMID:12136140.
- Song SY et al. (1983), J Mol Biol, 171, 225-228. Preliminary crystallographic studies of lobster D-glyceraldehyde-3-phosphate dehydrogenase and the modified enzyme carrying the fluorescent derivative. PMID:6655693.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys149(148)G(A) | proton shuttle (general acid/base) |
| His176(175)G(A) | proton shuttle (general acid/base) |
| Cys149(148)G(A) | covalent catalysis |