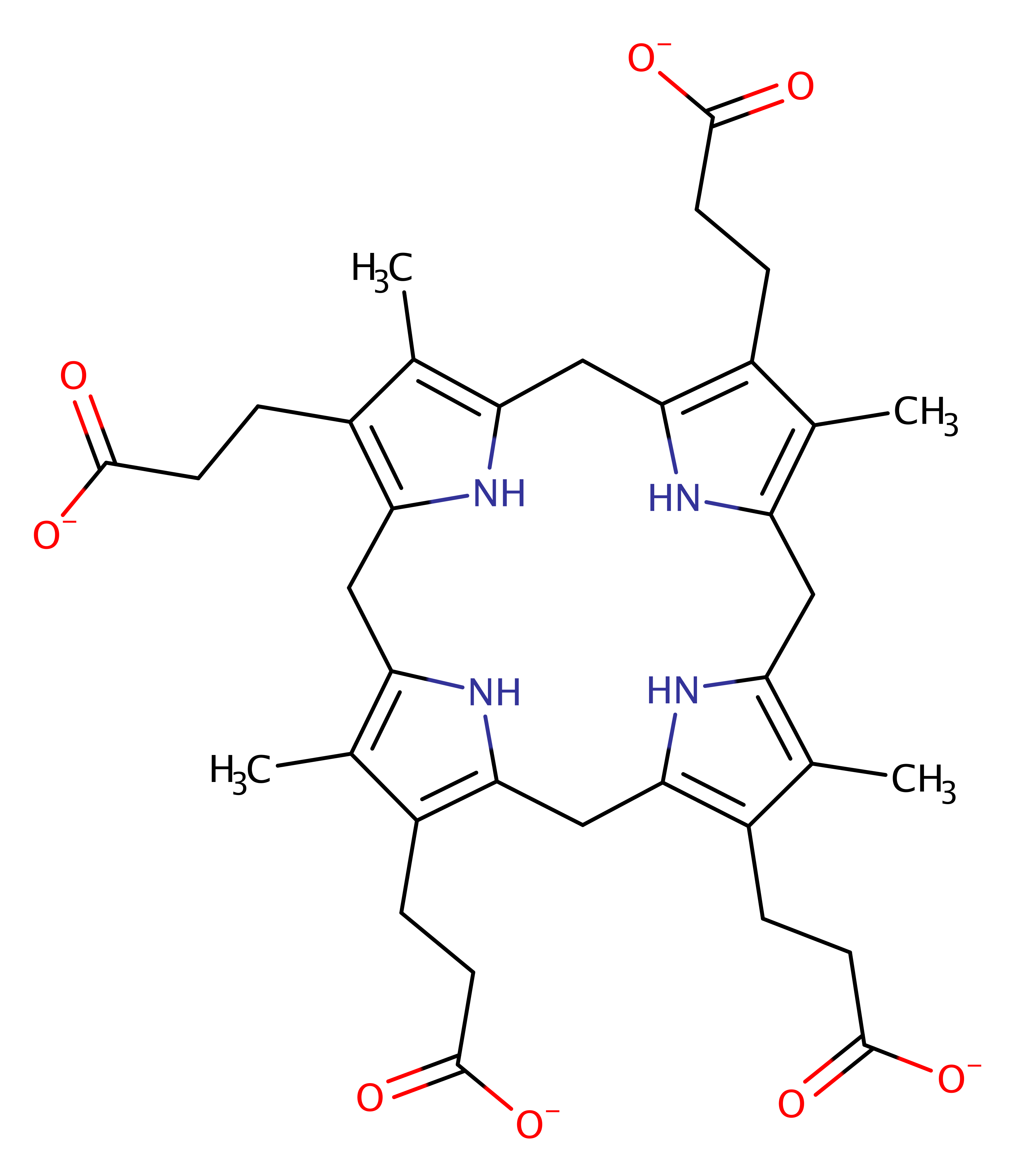
Uroporphyrinogen decarboxylase
Uroporphyrinogen decarboxylase (HemE) catalyses the fifth step in heme biosynthesis by converting uroporphyrinogen III to coproporphyrinogen III by decarboxylating the four acetate side chains of the substrate.
Reference Protein and Structure
- Sequence
-
P06132
 (4.1.1.37)
(4.1.1.37)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1uro
- UROPORPHYRINOGEN DECARBOXYLASE
(1.8 Å)



- Catalytic CATH Domains
-
3.20.20.210
 (see all for 1uro)
(see all for 1uro)
Enzyme Reaction (EC:4.1.1.37)
Enzyme Mechanism
Introduction
The first step in the proposed mechanism is a proton transfer from an acid (Arg37) to the C2 centre of ring D. Computational studies suggest that this first step is rate limiting. The next step is the decarboxylation of the acetate moiety. The final catalytic step in the overall mechanism is regeneration of the protonated Arg37 residue by transfer of a proton from the substrate's --C2H2-- group to its neutral guanidino group with concurrent proton transfer from the guanidinium group of Arg50 to the C3′ centre of the substrate.
Catalytic Residues Roles
| UniProt | PDB* (1uro) | ||
| Arg37 | Arg37A | Acts as a general acid/base. | proton shuttle (general acid/base) |
| Arg50, Arg41 | Arg50A, Arg41A | There is some debate as to the exact roles of these two argenines. It has been suggested that one acts as a general acid/base and the other helps stabilise the reactive intermediates and transition states formed during the course of the reaction. We have annotated the roles based on the primary reference in which Arg41 is stabilising and Arg50 is the general acid/base. | proton shuttle (general acid/base) |
| Asp86 | Asp86A | A reduced Asp86---pyrrole carbocation interaction causes a destabilisation of the carbocation, thus enhancing the feasibility of decarboxylation. A further role of Asp86 is to enhance the basicity of C3′ in key mechanistic intermediates. It is also thought to act as a transition state stabiliser. | modifies pKa, enhance reactivity, transition state stabiliser |
| Tyr164 | Tyr164A | Whilst not essential for catalysis, it is thought that this residue helps drive the decarboxylation step through electrostatic interactions. | electrostatic stabiliser |
Chemical Components
References
- Bushnell EA et al. (2011), J Comput Chem, 32, 822-834. The first branching point in porphyrin biosynthesis: a systematic docking, molecular dynamics and quantum mechanical/molecular mechanical study of substrate binding and mechanism of uroporphyrinogen-III decarboxylase. DOI:10.1002/jcc.21661. PMID:20941734.
- Silva PJ et al. (2010), J Phys Chem B, 114, 8994-9001. A tale of two acids: when arginine is a more appropriate acid than H3O+. DOI:10.1021/jp100961s. PMID:20553007.
- Phillips JD et al. (2009), J Mol Biol, 389, 306-314. Substrate shuttling between active sites of uroporphyrinogen decarboxylase is not required to generate coproporphyrinogen. DOI:10.1016/j.jmb.2009.04.013. PMID:19362562.
- Fan J et al. (2007), J Bacteriol, 189, 3573-3580. Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis. DOI:10.1128/JB.01083-06. PMID:17122346.
- Phillips JD et al. (2003), EMBO J, 22, 6225-6233. Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase. DOI:10.1093/emboj/cdg606. PMID:14633982.
- Martins BM et al. (2001), J Biol Chem, 276, 44108-44116. Crystal Structure and Substrate Binding Modeling of the Uroporphyrinogen-III Decarboxylase from Nicotiana tabacum: IMPLICATIONS FOR THE CATALYTIC MECHANISM. DOI:10.1074/jbc.m104759200. PMID:11524417.
- Whitby FG et al. (1998), EMBO J, 17, 2463-2471. Crystal structure of human uroporphyrinogen decarboxylase. DOI:10.1093/emboj/17.9.2463. PMID:9564029.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp86A | modifies pKa, enhance reactivity |
| Arg37A | proton shuttle (general acid/base) |
| Arg50A | proton shuttle (general acid/base) |
| Tyr164A | electrostatic stabiliser |
| Asp86A | transition state stabiliser |
| Arg41A | electrostatic stabiliser |