Inorganic diphosphatase
Soluble inorganic pyrophosphate is present in all organisms, and is essential for cell growth. It provides an entropic pull for biosynthetic reactions involving nucleotide triphosphates by irreversibly hydrolysing the pyrophosphate product to orthophosphate. The phosphoryltransferase activity is an ancient one, and there are functional similarities between the active site of this enzyme and alkaline phosphatase and other divalent cation containing enzymes such as exonucleases and polymerases.
Reference Protein and Structure
- Sequence
-
P9WI55
 (3.6.1.1)
(3.6.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Mycobacterium tuberculosis H37Rv (Bacteria)

- PDB
-
4z71
- Crystal structure of inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with Mg ions
(1.85 Å)



- Catalytic CATH Domains
-
3.90.80.10
 (see all for 4z71)
(see all for 4z71)
- Cofactors
- Magnesium(2+) (2)
Enzyme Reaction (EC:3.6.1.1)
Enzyme Mechanism
Introduction
This reaction begins with the coordination of three divalent metal ions into the active site as well as a water molecule from the aqueous solution. The metal ions coordinate a diphosphate into the active site while Asp89 deprotonates a water molecule. The resulting hydroxide attacks the proximal phosphate. This, in combination with the metals dissociating from the active site results in the release of two phosphate molecules. His21 and His86 are responsible for divalent metal selectivity and are major features distinguishing the Mtb PPiase from other characterised PPiases but aren't shown in this mechanism because they don't directly contribute to the catalysis reaction.
Catalytic Residues Roles
| UniProt | PDB* (4z71) | ||
| Asp89 | Asp89(98)A | Thought to act as a general acid/base. It deprotonates a water molecule that reacts with the substrate. | activator, metal ligand, proton acceptor |
| Asp57, Asp89, Asp52, Glu8, Asp84 | Asp57(66)A, Asp89(98)A, Asp52(61)A, Glu8(17)A, Asp84(93)A | Metal ligands. | metal ligand |
Chemical Components
overall product formed, proton transfer, bimolecular nucleophilic substitution, overall reactant used, coordination to a metal ionReferences
- Pratt AC et al. (2015), J Struct Biol, 192, 76-87. Structural and computational dissection of the catalytic mechanism of the inorganic pyrophosphatase from Mycobacterium tuberculosis. DOI:10.1016/j.jsb.2015.08.010. PMID:26296329.
- Pang AH et al. (2016), ACS Chem Biol, 11, 3084-3092. Discovery of Allosteric and Selective Inhibitors of Inorganic Pyrophosphatase from Mycobacterium tuberculosis. DOI:10.1021/acschembio.6b00510. PMID:27622287.
- Rodina EV et al. (2008), Biochemistry (Mosc), 73, 897-905. Metal cofactors play a dual role in Mycobacterium tuberculosis inorganic pyrophosphatase. DOI:10.1134/s0006297908080075. PMID:18774936.
- Halonen P et al. (2005), Biochemistry, 44, 4004-4010. Effects of Active Site Mutations on the Metal Binding Affinity, Catalytic Competence, and Stability of the Family II Pyrophosphatase fromBacillus subtilis†. DOI:10.1021/bi047926u. PMID:15751976.
- Fabrichniy IP et al. (2004), Biochemistry, 43, 14403-14411. Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion†,‡. DOI:10.1021/bi0484973. PMID:15533045.
- Zyryanov AB et al. (2004), Biochemistry, 43, 1065-1074. Rates of Elementary Catalytic Steps for Different Metal Forms of the Family II Pyrophosphatase fromStreptococcus gordonii†. DOI:10.1021/bi0357513. PMID:14744152.
- Zyryanov AB et al. (2004), Biochemistry, 43, 14395-14402. Site-specific effects of zinc on the activity of family II pyrophosphatase. DOI:10.1021/bi048470j. PMID:15533044.
- Shizawa N et al. (2001), Eur J Biochem, 268, 5771-5775. Directed mutagenesis studies of the C-terminal fingerprint region ofBacillus subtilispyrophosphatase. DOI:10.1046/j.0014-2956.2001.02513.x. PMID:11722562.
- Heikinheimo P et al. (1996), Eur J Biochem, 239, 138-143. A site-directed mutagenesis study of Saccharomyces cerevisiae pyrophosphatase. Functional conservation of the active site of soluble inorganic pyrophosphatases. PMID:8706698.
- Harutyunyan EH et al. (1996), Eur J Biochem, 239, 220-228. X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate. PMID:8706712.
- Raznikov AV et al. (1992), FEBS Lett, 308, 62-64. Tyrosine-89 is important for enzymatic activity of S. cerevisiae inorganic pyrophosphatase. PMID:1322842.

Step 1. A divalent metal ion M3 binds to the PPiase, which already contains the divalent metal ion M1 in its active site. The presence of the two metal ions enable the binding of PPi together with a third divalent metal ion: M2.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp89(98)A | metal ligand |
| Asp52(61)A | metal ligand |
| Asp57(66)A | metal ligand |
Chemical Components

Step 2. The binding events are followed by the deprotonation of a water molecule by Asp89 and subsequent attack on the adjacent phosphate group of PPi by the resulting hydroxide, leading to the formation of the products.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp52(61)A | metal ligand |
| Glu8(17)A | metal ligand |
| Asp57(66)A | metal ligand |
| Asp84(93)A | metal ligand |
| Asp89(98)A | proton acceptor, metal ligand, activator |
Chemical Components
overall product formed, proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, coordination to a metal ion
Step 3. After catalysis, ion M3 leaves the active site, followed by both phosphates and M2. Also, Asp89 is deprotonated (here inferred to be by a water molecule from solvent), thereby restoring the initial state of enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp89(98)A | metal ligand |
| Asp52(61)A | metal ligand |
| Glu8(17)A | metal ligand |
| Asp57(66)A | metal ligand |
| Asp84(93)A | metal ligand |
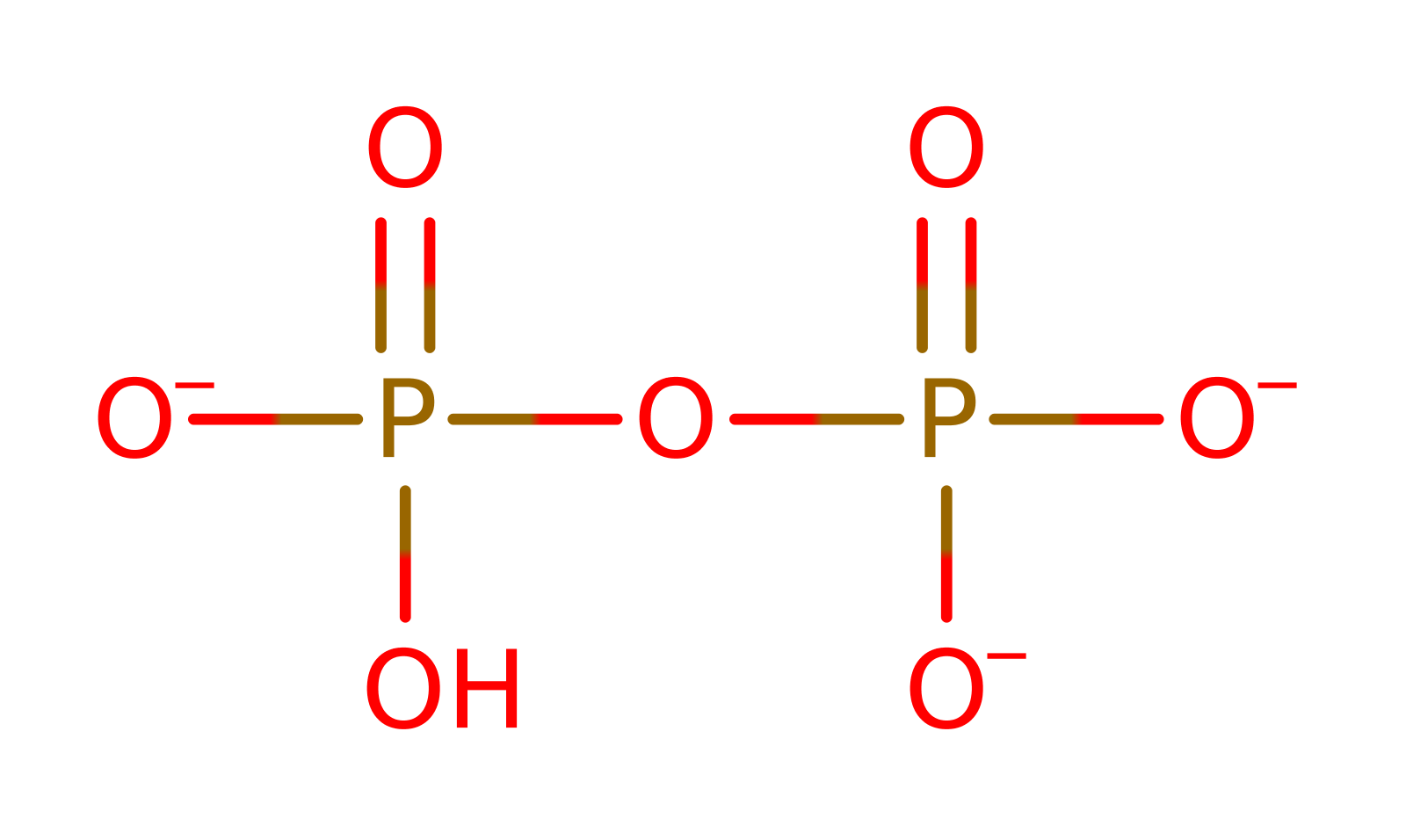



 Download:
Download: