Hydroxymethylglutaryl-CoA reductase (NADPH)
Hydroxymethylglutaryl-CoA reductase catalyses the committed step in the biosynthesis of isoprenoids, compounds involved in diverse cellular functions such as sterol synthesis and growth control. The precursor of such products is melvalonic acid and concentrations of mevalonate are tightly controlled within cells. This is achieved through the activity of 3-hydroxy-3-methylglutaryl-CoA reductase which catalyses the four electron reduction reduction of 3-hydroxy-3-methylglutaryl-CoA to mevalonate. The activity of 3-hydroxy-3-methylglutaryl-CoA reductase is controlled physiologically through synthesis, degradation and phosphorylation as well as clinically, being the target of several high cholesterol drugs.
Reference Protein and Structure
- Sequence
-
P04035
 (1.1.1.34)
(1.1.1.34)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1dqa
- COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG, COA, AND NADP+
(2.0 Å)



- Catalytic CATH Domains
-
3.90.770.10
 3.30.70.420
3.30.70.420  (see all for 1dqa)
(see all for 1dqa)
Enzyme Reaction (EC:1.1.1.34)
Enzyme Mechanism
Introduction
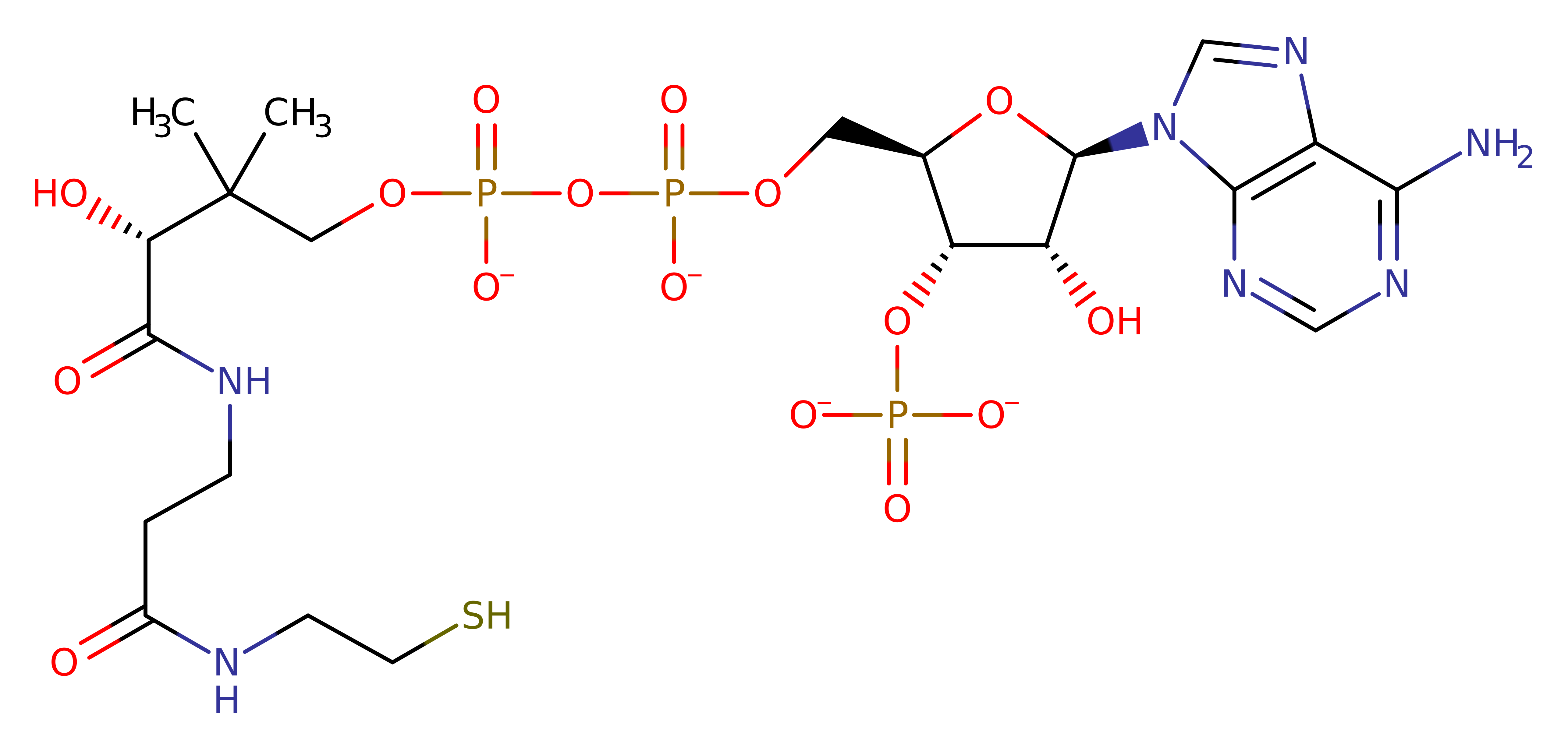
The first reduction step to mevaldyl-CoA, with concomittant deprotonation of Glu559. In the second step Glu559 deprotonates the hydroxide formed in step one, and the eliminated thiolate deprotonates His866. In the third step several residues are involved in the reduction of mevaldehyde. Glu559 is thought to be the proton donor for mevaldehyde.
Catalytic Residues Roles
| UniProt | PDB* (1dqa) | ||
| Glu559, His866 | Glu559(138)B, His866(445)B | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Lys691, Asp767 | Lys691(270)A, Asp767(346)A | Acts to stabilise the reactive intermediates in the active site. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
aromatic unimolecular elimination by the conjugate base, hydride transfer, bimolecular nucleophilic addition, proton transfer, bimolecular elimination, native state of enzyme regenerated, inferred reaction stepReferences
- Haines BE et al. (2012), Biochemistry, 51, 7983-7995. Molecular Modeling of the Reaction Pathway and Hydride Transfer Reactions of HMG-CoA Reductase. DOI:10.1021/bi3008593. PMID:22971202.
- Oliveira EF et al. (2016), Catal Sci Technol, 6, 7172-7185. QM/MM study of the mechanism of reduction of 3-hydroxy-3-methylglutaryl coenzyme A catalyzed by human HMG-CoA reductase. DOI:10.1039/c6cy00356g.
- Haines BE et al. (2013), Acc Chem Res, 46, 2416-2426. The Increasingly Complex Mechanism of HMG-CoA Reductase. DOI:10.1021/ar3003267. PMID:23898905.

Step 1. NADP eliminates a hydride, which adds to the carbonyl carbon adjacent to the CoA moiety.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys691(270)A | hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu559(138)B | proton donor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, hydride transfer, ingold: bimolecular nucleophilic addition, proton transfer
Step 2. The oxyanion collapses, eliminating CoA, which deprotonates His866B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His866(445)B | hydrogen bond donor |
| Lys691(270)A | electrostatic stabiliser, hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor, electrostatic stabiliser |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu559(138)B | proton acceptor |
| His866(445)B | proton donor |
Chemical Components
proton transfer, ingold: bimolecular elimination
Step 3. A second molecule of NADP eliminates a hydride, which adds to the terminal carbonyl carbon, which deprotonates Glu559B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys691(270)A | hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu559(138)B | proton donor |
Chemical Components
proton transfer, hydride transfer, ingold: bimolecular nucleophilic addition, ingold: aromatic unimolecular elimination by the conjugate baseCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His866(445)B | hydrogen bond acceptor |
| Glu559(138)B | hydrogen bond acceptor |
| Asp767(346)A | hydrogen bond acceptor, activator |
| His866(445)B | proton acceptor |
| Glu559(138)B | proton acceptor |
Chemical Components
proton transfer, native state of enzyme regenerated, inferred reaction stepIntroduction
The first reduction step to mevaldyl-CoA leaves a negatively charged oxygen which is stabilised by Lys691. In the second step His866 is proposed as a proton donor to the thioanion. In the third step several residues are involved in the reduction of mevaldehyde. Glu559, its pKa raised by the close proximity of Asp767, is thought to be the proton donor for mevaldehyde.
Catalytic Residues Roles
| UniProt | PDB* (1dqa) | ||
| Glu559, His866 | Glu559(138)B, His866(445)B | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Lys691, Asp767 | Lys691(270)A, Asp767(346)A | Acts as an electrostatic stabiliser. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
aromatic unimolecular elimination by the conjugate base, hydride transfer, bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate base, native state of enzyme regenerated, inferred reaction stepReferences
- Istvan ES et al. (2000), EMBO J, 19, 819-830. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. DOI:10.1093/emboj/19.5.819. PMID:10698924.
- Oliveira EF et al. (2016), Catal Sci Technol, 6, 7172-7185. QM/MM study of the mechanism of reduction of 3-hydroxy-3-methylglutaryl coenzyme A catalyzed by human HMG-CoA reductase. DOI:10.1039/c6cy00356g.
- Leichner GS et al. (2009), Mol Biol Cell, 20, 3330-3341. Dislocation of HMG-CoA Reductase and Insig-1, Two Polytopic Endoplasmic Reticulum Proteins, En Route to Proteasomal Degradation. DOI:10.1091/mbc.e08-09-0953. PMID:19458199.
- Friesen JA et al. (2004), Genome Biol, 5, 248-. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. DOI:10.1186/gb-2004-5-11-248. PMID:15535874.
- Istvan ES et al. (2000), Biochim Biophys Acta, 1529, 9-18. The structure of the catalytic portion of human HMG-CoA reductase. DOI:10.1016/s1388-1981(00)00134-7. PMID:11111074.

Step 1. NADP eliminates a hydride, which adds to the carbonyl carbon adjacent to the CoA moiety.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys691(270)A | hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys691(270)A | electrostatic stabiliser |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, hydride transfer, ingold: bimolecular nucleophilic addition
Step 2. The oxyanion collapses, eliminating CoA, which deprotonates His866B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His866(445)B | hydrogen bond donor |
| Lys691(270)A | electrostatic stabiliser, hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor, electrostatic stabiliser |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| His866(445)B | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base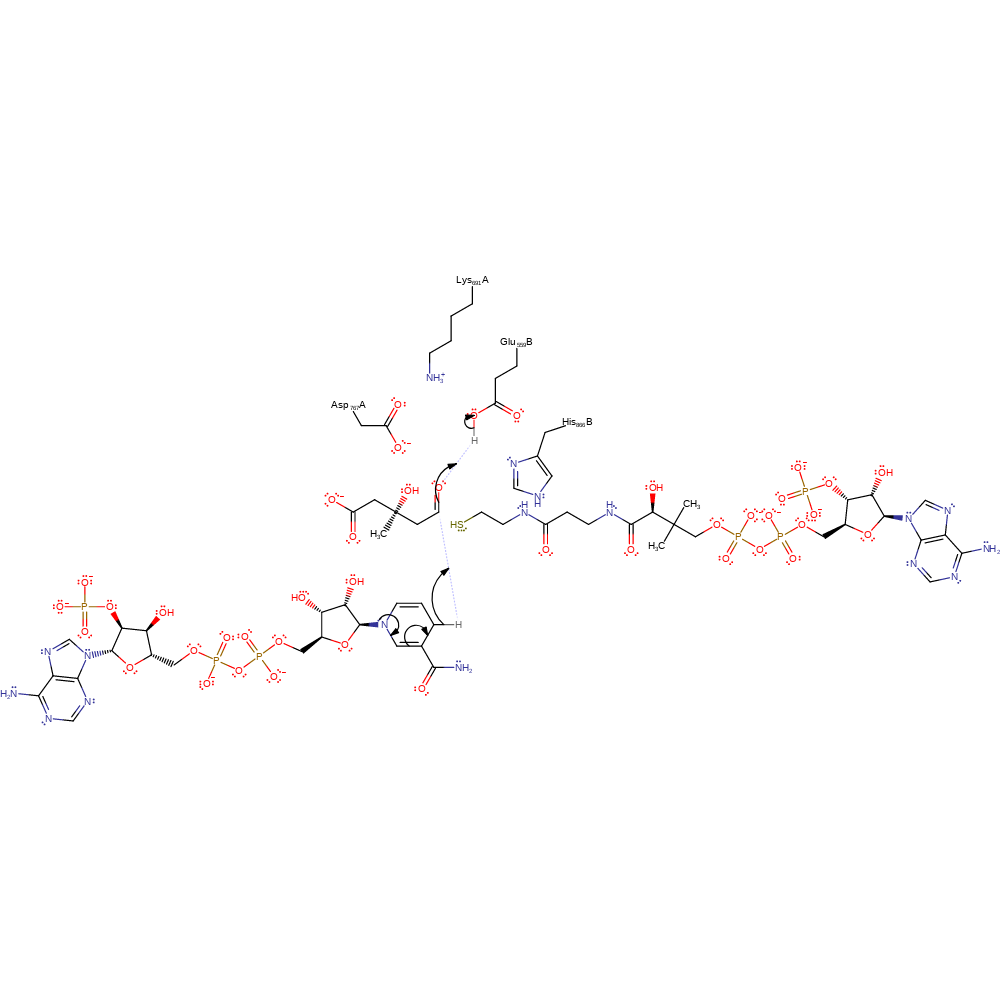
Step 3. A second molecule of NADP eliminates a hydride, which adds to the terminal carbonyl carbon, which deprotonates Glu559B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys691(270)A | hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys691(270)A | electrostatic stabiliser |
| Glu559(138)B | proton donor |
Chemical Components
proton transfer, hydride transfer, ingold: bimolecular nucleophilic addition, ingold: aromatic unimolecular elimination by the conjugate baseCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His866(445)B | hydrogen bond acceptor |
| Glu559(138)B | hydrogen bond acceptor |
| Asp767(346)A | hydrogen bond acceptor, activator |
| Glu559(138)B | proton acceptor |
| His866(445)B | proton acceptor |
Chemical Components
proton transfer, native state of enzyme regenerated, inferred reaction stepIntroduction
The first reduction step to mevaldyl-CoA leaves a negatively charged oxygen which is stabilised by Lys691. In the second step His866 is proposed as a proton donor to the thioanion. In the third step several residues are involved in the reduction of mevaldehyde. Lys691 is thought to be the proton donor for mevaldehyde.
Catalytic Residues Roles
| UniProt | PDB* (1dqa) | ||
| His866, Lys691 | His866(445)B, Lys691(270)A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu559, Asp767 | Glu559(138)B, Asp767(346)A | Acts to stabilise the reactive intermediates in the active site. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
aromatic unimolecular elimination by the conjugate base, hydride transfer, bimolecular nucleophilic addition, proton transfer, unimolecular elimination by the conjugate base, overall product formed, native state of enzyme regenerated, inferred reaction stepReferences
- Tabernero L et al. (1999), Proc Natl Acad Sci U S A, 96, 7167-7171. Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase. DOI:10.1073/pnas.96.13.7167.
- Oliveira EF et al. (2016), Catal Sci Technol, 6, 7172-7185. QM/MM study of the mechanism of reduction of 3-hydroxy-3-methylglutaryl coenzyme A catalyzed by human HMG-CoA reductase. DOI:10.1039/c6cy00356g.

Step 1. NADP eliminates a hydride, which adds to the carbonyl carbon adjacent to the CoA moiety.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys691(270)A | hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, hydride transfer, ingold: bimolecular nucleophilic addition
Step 2. The oxyanion collapses, eliminating CoA, which deprotonates His866B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His866(445)B | hydrogen bond donor |
| Lys691(270)A | electrostatic stabiliser, hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor, electrostatic stabiliser |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| His866(445)B | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, overall product formed
Step 3. A second molecule of NADP eliminates a hydride, which adds to the terminal carbonyl carbon, which deprotonates Lys691.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys691(270)A | hydrogen bond donor |
| Glu559(138)B | hydrogen bond donor |
| Asp767(346)A | hydrogen bond acceptor, electrostatic stabiliser |
| Glu559(138)B | electrostatic stabiliser |
| Lys691(270)A | proton donor |
Chemical Components
proton transfer, hydride transfer, ingold: bimolecular nucleophilic addition, ingold: aromatic unimolecular elimination by the conjugate base
Step 4. Lys691 and His866B deprotonate water in an inferred return step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His866(445)B | hydrogen bond acceptor |
| Glu559(138)B | hydrogen bond acceptor |
| Asp767(346)A | hydrogen bond acceptor, activator |
| His866(445)B | proton acceptor |
| Lys691(270)A | proton acceptor |






 Download:
Download: 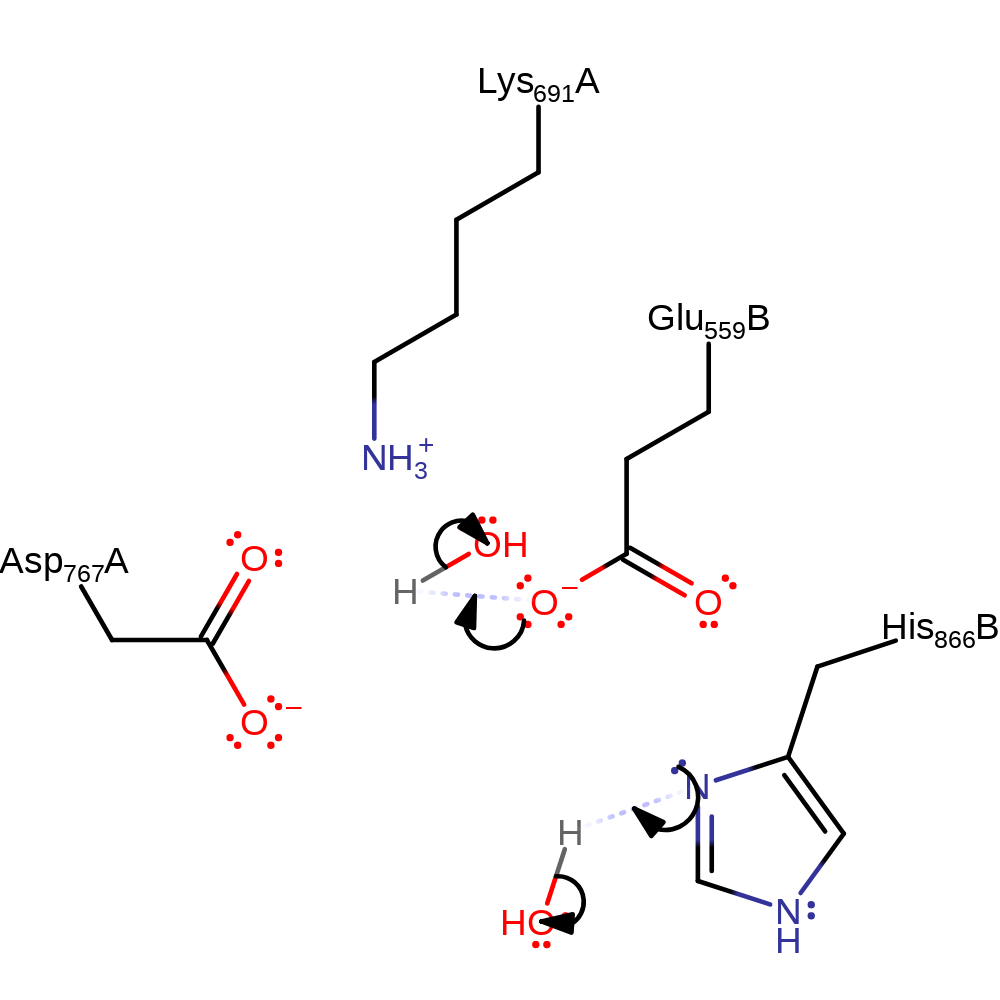
 Download:
Download:  Download:
Download: