Nicotinate-nucleotide pyrophosphorylase (carboxylating) (type II)
This is a type II Quinolinic acid phosphoribosyltransferase (QAPRTase).
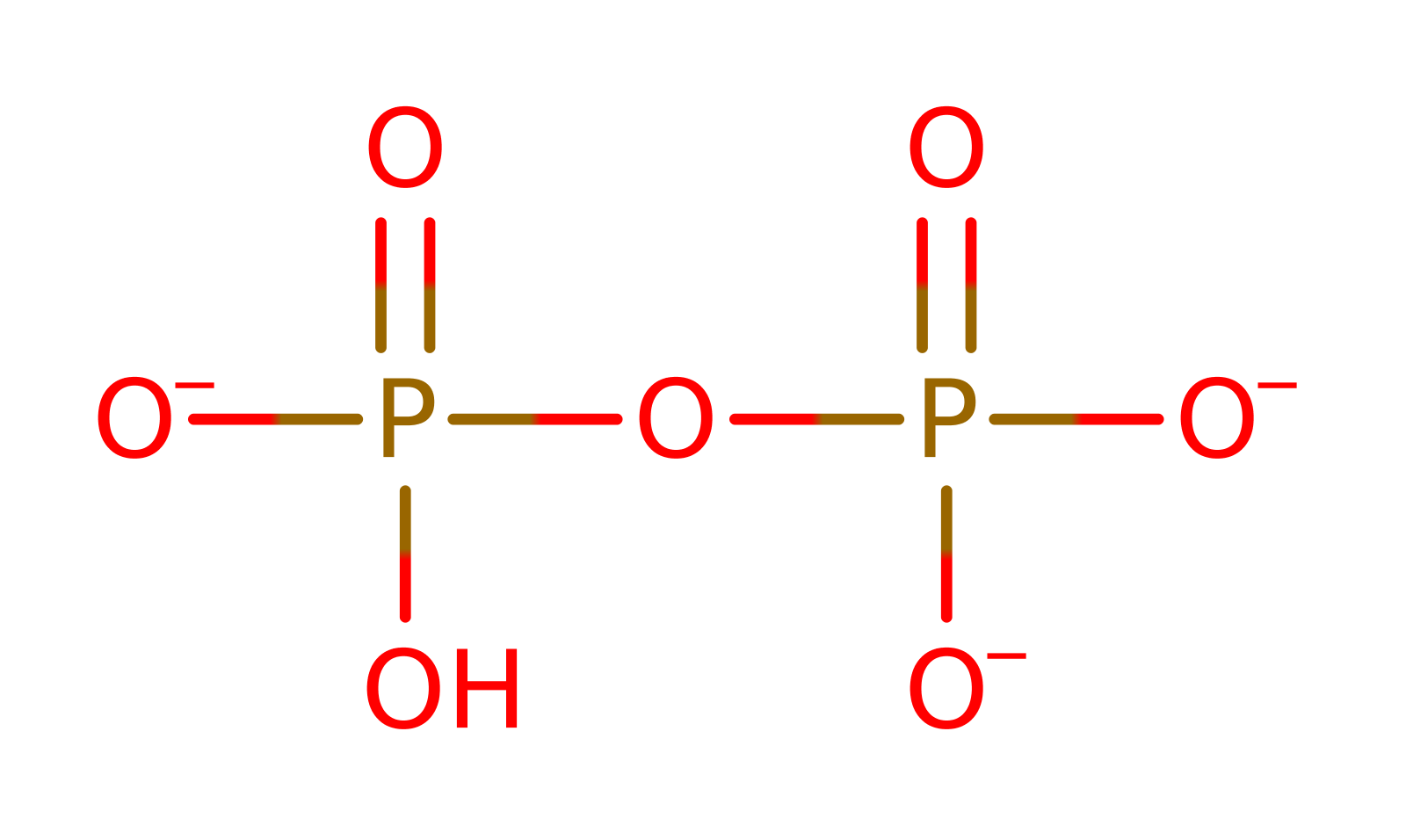
This protein is required for the de novo biosynthesis of NAD in both prokaryotes and eukaryotes. The enzyme catalyses the reaction between quinolinic acid (QA) and 5-phosphoribosyl-1-pyrophosphate (PRPP), to yield nicotinic acid mononucleotide (NAMN), pyrophosphate and CO2, the latter resulting from decarboxylation at position 2 of the quinolinate ring.
QAPRTase has been grouped with nine other enzymes, (phosphoribosyltransferases, PRTases) that catalyse chemically similar phosphoribosyl transfer reactions using the substrate PRPP. The PRTases are involved in de novo and salvage reactions of nucleotide synthesis, as well as in histidine and tryptophan biosynthesis. Type II enzymes lack the type I PRPP-binding motif and have TIM barrel-like structure [PMID:9016724].
Reference Protein and Structure
- Sequence
-
P43619
 (2.4.2.19)
(2.4.2.19)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
3c2e
- Crystal structure at 1.9A of the apo quinolinate phosphoribosyl transferase (BNA6) from Saccharomyces cerevisiae
(1.9 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 3c2e)
(see all for 3c2e)
Enzyme Reaction (EC:2.4.2.19)
Enzyme Mechanism
Introduction
Phosphoribosyl transfer has been proposed to proceed via a unimolecular nucleophilic substitution (SN1 reaction) involving an oxycarbonium-like intermediate. In the first step of the reaction the sugar ring eliminates the diphosphate group. In the second step, the carboxylic acid group forms carbon dioxide, which initiates the nucleophilic addition of the pyridine nitrogen to the oxycarbonium-like intermediate to form the final products.
Catalytic Residues Roles
| UniProt | PDB* (3c2e) | ||
| Lys144 | Lys144(143)A | Helps stabilise the reactive intermediates and transition states formed during the course of the reaction. | electrostatic stabiliser |
Chemical Components
References
- di Luccio E et al. (2008), Biochemistry, 47, 4039-4050. Comprehensive X-ray Structural Studies of the Quinolinate Phosphoribosyl Transferase (BNA6) fromSaccharomyces cerevisiae‡. DOI:10.1021/bi7020475. PMID:18321072.
- Eads JC et al. (1997), Structure, 5, 47-58. A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase. DOI:10.1016/s0969-2126(97)00165-2. PMID:9016724.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys144(143)A | electrostatic stabiliser |