Phosphohistidine phosphatase
This eukaryotic enzyme dephosphorylates phosphorylated histidine residues within proteins and peptides. The enzyme acts on phosphate groups attached to both the pros- and tele-nitrogen atoms, but the pros- position is somewhat preferred (by a factor of two at the most) [PMID:19836471]. The substrate specificity depends on the amino acid sequence or structural context of the phosphohistidine in a phosphoprotein. The enzyme is also active on free phosphoramidate [PMID:12383260, PMID:19836471] and peptide-bound phospholysine [PMID:25574816].
Reference Protein and Structure
- Sequence
-
Q9NRX4
 (3.9.1.3)
(3.9.1.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
2hw4
- Crystal structure of human phosphohistidine phosphatase
(1.9 Å)



- Catalytic CATH Domains
-
3.50.20.20
 (see all for 2hw4)
(see all for 2hw4)
Enzyme Reaction (EC:3.9.1.3)
Enzyme Mechanism
Introduction
The substrate binds to the protein. His53 acts as a general base to activate a water molecule, which in turn functions as a nucleophile to attack the phosphate of the substrate. The phosphate is eliminated and the substrate histidine abstracts a proton from His53 (inferred).
Catalytic Residues Roles
| UniProt | PDB* (2hw4) | ||
| His53 | His53(72)A | Acts as a general acid/base. | proton acceptor, proton donor |
| Ala54 (main-N), Ser94, Ala96 (main-N), Arg78 | Ala54(73)A (main-N), Ser94(113)A, Ala96(115)A (main-N), Arg78(97)A | Form a positively charged phosphate binding site which helps stabilise the reactive intermediates and transition states formed during the course of the reaction. | electrostatic stabiliser |
| Lys21 | Lys21(40)A | Lys21 helps stabilise the reactive intermediates and transition states formed during the course of the reaction. The amine group of Lys21 is also close to the imidazole ring of His53, and the positive charge may also be a reason for the reduced pKa of the imidazole ring | modifies pKa, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall product formed, overall reactant used, inferred reaction step, native state of enzyme regeneratedReferences
- Gong W et al. (2009), Biochem J, 418, 337-344. Solution structure and catalytic mechanism of human protein histidine phosphatase 1. DOI:10.1042/BJ20081571. PMID:18991813.
- Ek P et al. (2015), Ups J Med Sci, 120, 20-27. Phosphohistidine phosphatase 1 (PHPT1) also dephosphorylates phospholysine of chemically phosphorylated histone H1 and polylysine. DOI:10.3109/03009734.2014.996720. PMID:25574816.
- Attwood PV et al. (2010), Biochim Biophys Acta, 1804, 199-205. Chemical phosphorylation of histidine-containing peptides based on the sequence of histone H4 and their dephosphorylation by protein histidine phosphatase. DOI:10.1016/j.bbapap.2009.10.007. PMID:19836471.
- Busam RD et al. (2006), J Biol Chem, 281, 33830-33834. First structure of a eukaryotic phosphohistidine phosphatase. DOI:10.1074/jbc.C600231200. PMID:16990267.
- Ma R et al. (2005), Biochem Biophys Res Commun, 337, 887-891. Mutational study of human phosphohistidine phosphatase: effect on enzymatic activity. DOI:10.1016/j.bbrc.2005.09.134. PMID:16219293.
- Ek P et al. (2002), Eur J Biochem, 269, 5016-5023. Identification and characterization of a mammalian 14-kDa phosphohistidine phosphatase. PMID:12383260.

Step 1. His53 abstracts a proton from the nucleophilic water, which attacks the phosphate, eliminating the protein histidine intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys21(40)A | electrostatic stabiliser |
| Ala54(73)A (main-N) | electrostatic stabiliser |
| Arg78(97)A | electrostatic stabiliser |
| Ser94(113)A | electrostatic stabiliser |
| Ala96(115)A (main-N) | electrostatic stabiliser |
| Lys21(40)A | modifies pKa |
| His53(72)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall product formed, overall reactant used
Step 2. Inferred step in which the protein histidine intermediate abstracts a proton from the catalytic histidine residue to regenerate the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys21(40)A | modifies pKa |
| His53(72)A | proton donor |


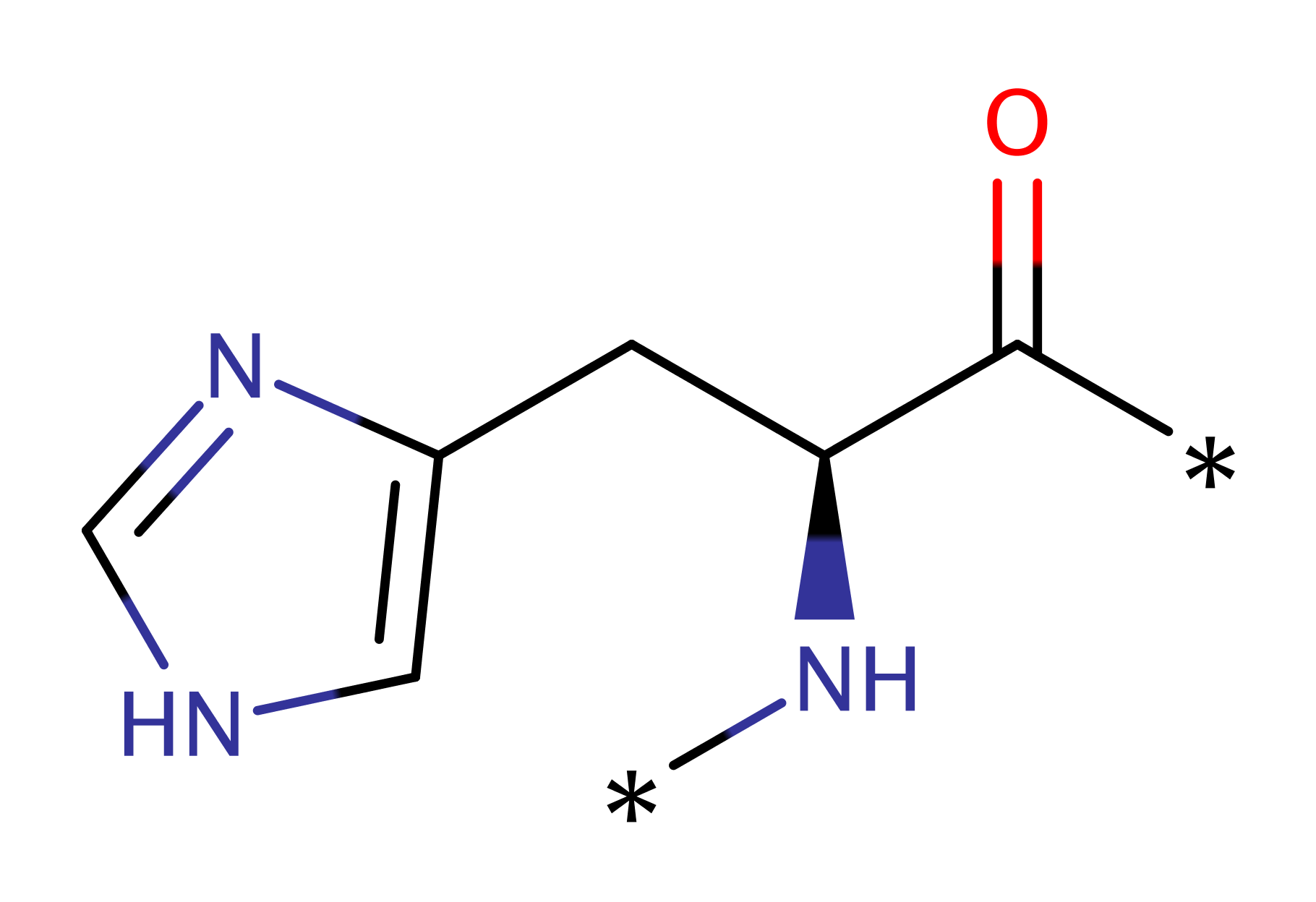

 Download:
Download: