Cytidine deaminase
Cytosine deaminase (CD) catalyses the deamination of cytosine, producing uracil. This enzyme is present in prokaryotes and fungi (but not multicellular eukaryotes) and is an important member of the pyrimidine salvage pathway in those organisms. The enzyme is of widespread interest both for antimicrobial drug design and for gene therapy applications against tumours.
Reference Protein and Structure
- Sequence
-
P0ABF6
 (3.5.4.5)
(3.5.4.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1ctt
- TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE
(2.2 Å)



- Catalytic CATH Domains
-
3.40.140.10
 (see all for 1ctt)
(see all for 1ctt)
- Cofactors
- Zinc(2+) (1) Metal MACiE
Enzyme Reaction (EC:3.5.4.5)
Enzyme Mechanism
Introduction
The full catalytic cycle is composed of three major steps, including the substrate (cytosine) binding, the release of ammonia and the departure of the product (uracil) from the active site.
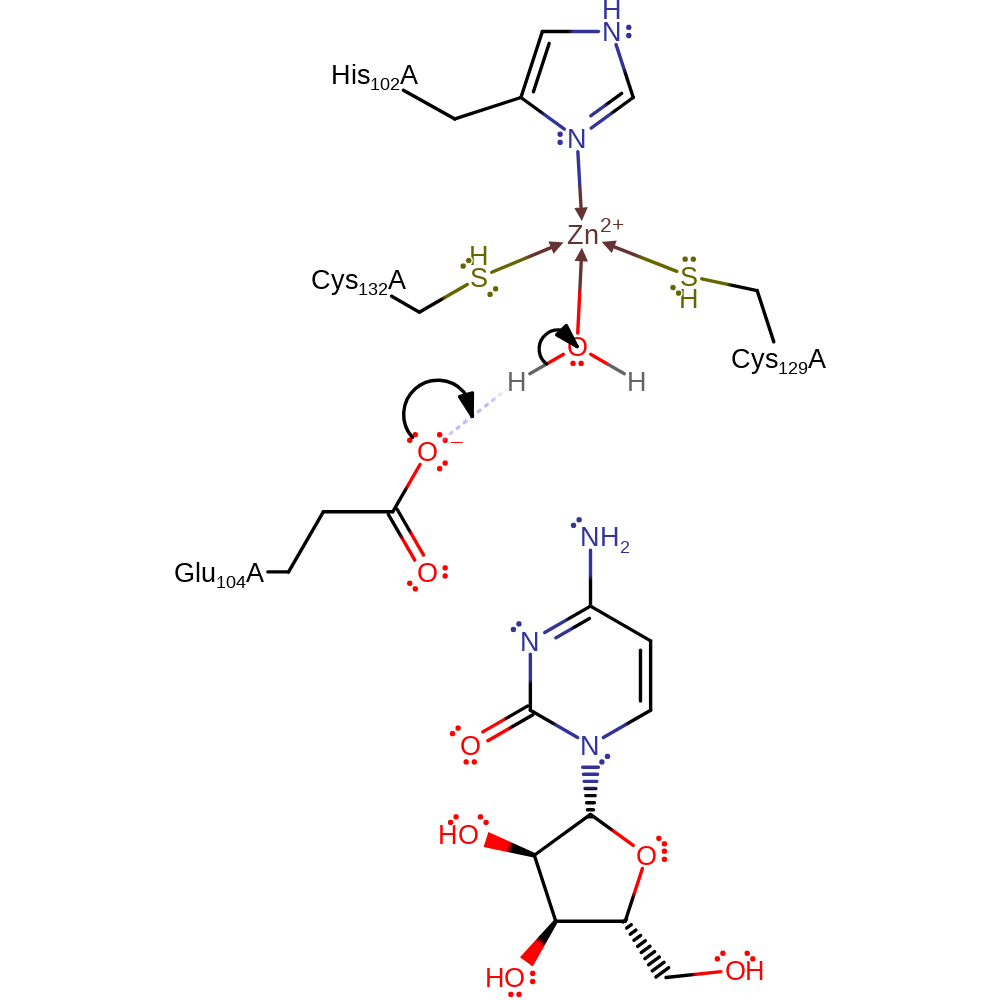
The catalytic zinc ion is bound to His102, Cys129, Cys132, and a catalytic water. The positive charge of the metal ion polarises the O-H bond in water, increasing its acidity towards the close proximity Glu104. Thus, the first step involved the proton transfer from the zinc-bound water to Glu104, the rotation of the carboxyl group of Glu104 and the proton transfer from the protonated Glu104 to cytosine. The subsequent nucleophilic attack at the C2 of the substrate by the resulting hydroxide anion leads to a tetrahedral intermediate. The second step concerns the formation of both ammonia and uracil. Once again this steps involves the proton transfers from the zinc-bound hydroxide anion to Glu104 and from Glu104 to the amine group which are stepwise rather than synchronous as in the first step. This step forms ammonia and uracil. Uracil remains strongly bound to the zinc metal. The final step consists of several proton transfers shuttled by Glu104 and one nucleophilic attack. One water molecule, which replaces the ammonia from the second step, heterolytically splits with its proton going to Glu104 and the hydroxide anion synchronously attacking C2 of uracil.
Catalytic Residues Roles
| UniProt | PDB* (1ctt) | ||
| Cys132 | Cys132A | The Zn-Cys132 bond has a unique capacity to buffer changes in net negative charge across the zinc coordination sphere. This compensates for the developing negative charge on the substrate's 4-OH group as the reaction proceeds through the alkoxide character transition state. This valence-buffering may also be crucial in lowering the activation barrier associated with proton transfer steps in the hydration of the 3-4 N,C double bond. | metal ligand |
| Glu104 | Glu104A | The residue acts as a general base to the hydrolytic water molecule held in the Zn coordination sphere, which then attacks the substrate in an addition reaction with concurrent deprotonation of Glu104. The resulting carboxylate deprotonates the substrate hydroxyl, resulting in the loss of ammonia through a E1cb mechanism with abstraction of the Glu104 proton. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| His102, Cys129 | His102A, Cys129A | Form part of the zinc binding site. | metal ligand |
Chemical Components
proton transfer, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, deamination, native state of enzyme regeneratedReferences
- Zhang X et al. (2016), J Comput Chem, 37, 1163-1174. Combined QM(DFT)/MM molecular dynamics simulations of the deamination of cytosine by yeast cytosine deaminase (yCD). DOI:10.1002/jcc.24306. PMID:26813441.
- Hall RS et al. (2011), Biochemistry, 50, 5077-5085. Three-Dimensional Structure and Catalytic Mechanism of Cytosine Deaminase. DOI:10.1021/bi200483k. PMID:21545144.
- Xiang S et al. (1997), Biochemistry, 36, 4768-4774. The Structure of the Cytidine Deaminase−Product Complex Provides Evidence for Efficient Proton Transfer and Ground-State Destabilization†,‡. DOI:10.1021/bi963091e. PMID:9125497.
- Xiang S et al. (1996), Biochemistry, 35, 1335-1341. Cytidine Deaminase Complexed to 3-Deazacytidine: A “Valence Buffer” in Zinc Enzyme Catalysis†. DOI:10.1021/bi9525583. PMID:8634261.
- Betts L et al. (1994), J Mol Biol, 235, 635-656. Cytidine Deaminase. The 2·3 Å Crystal Structure of an Enzyme: Transition-state Analog Complex. DOI:10.1006/jmbi.1994.1018. PMID:8289286.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu104A | hydrogen bond acceptor |
| His102A | metal ligand |
| Cys132A | metal ligand |
| Cys129A | metal ligand |
| Glu104A | proton acceptor |
Chemical Components
proton transfer
Step 2. This step may well occur in a concerted manner with step 1. Glu104 rotates in the active site and donates its proton to the cytidine substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu104A | hydrogen bond donor |
| His102A | metal ligand |
| Cys132A | metal ligand |
| Cys129A | metal ligand |
| Glu104A | proton donor |
Chemical Components
proton transfer
Step 3. Activated water initiates a nucleophilic attack on the substrate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu104A | hydrogen bond acceptor |
| His102A | metal ligand |
| Cys132A | metal ligand |
| Cys129A | metal ligand |
| Glu104A | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic additionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His102A | metal ligand |
| Cys132A | metal ligand |
| Cys129A | metal ligand |
| Glu104A | proton acceptor |
Chemical Components
proton transfer
Step 5. The oxyanion collapses, eliminating ammonia with concomitant deprotonation of Glu104. Water displaces the product(s) from the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His102A | metal ligand |
| Cys132A | metal ligand |
| Cys129A | metal ligand |
| Glu104A | proton donor |




 Download:
Download: