Tryptophan 7-halogenase
Tryptophan 7-halogenase catalyses the chlorination of free tryptophan to 7-chlorotryptophan, which is the first step in the synthesis of the antibiotic pyrrolnitrin. Enzyme-catalysed chlorination of organic compounds in the laboratory remains a challenge, thus understanding the reaction mechanism of tryptophan 7-halogenase is important.
Reference Protein and Structure
- Sequence
-
P95480
 (1.14.19.9)
(1.14.19.9)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas fluorescens (Bacteria)

- PDB
-
2ar8
- The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination
(2.2 Å)



- Catalytic CATH Domains
-
3.50.50.60
 (see all for 2ar8)
(see all for 2ar8)
- Cofactors
- Nadp zwitterion (1)
Enzyme Reaction (EC:1.14.19.9)
Enzyme Mechanism
Introduction
In the FAD binding site, FADH2 reacts with molecular oxygen and subsequently with chlorine to form hypochlorous acid (HOCl). HOCl moves through a tunnel in the enzyme and becomes activated (Lys79) in order to chlorinate the substrate in the tryptophan binding site. The chlorine atom is inserted forming an arenium ion (Wheland intermediate). Proton removal of this reactive intermediate by Glu346 leads to the formation of the 7-chlorotryptophan product. The mechanism is regioselective. An alternative is the electrophilic attack performed by a preliminarily formed Lys79-bound chloramine. However, chloramines are milder chlorinating agents than HOCl and not sufficient for the reaction.
Catalytic Residues Roles
| UniProt | PDB* (2ar8) | ||
| Lys79 | Lys79A | Activates HOCl for electrophilic substitution on the tryptophan substrate. | activator, hydrogen bond donor, proton donor |
| Glu346 | Glu346A | Deprotonates Wheland intermediate forming the product. | proton acceptor |
Chemical Components
inferred reaction step, hydride transfer, cofactor used, electron transfer, proton transfer, radical formation, intermediate formation, colligation, radical termination, bimolecular nucleophilic substitution, dehydration, aromatic bimolecular electrophilic addition, aromatic bimolecular elimination, overall product formed, rate-determining stepReferences
- Karabencheva-Christova TG et al. (2017), Sci Rep, 7, 17395-. Mechanistic Insights into the Reaction of Chlorination of Tryptophan Catalyzed by Tryptophan 7-Halogenase. DOI:10.1038/s41598-017-17789-x. PMID:29234124.
- Romero E et al. (2018), Chem Rev, 118, 1742-1769. Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. DOI:10.1021/acs.chemrev.7b00650. PMID:29323892.
- Agarwal V et al. (2017), Chem Rev, 117, 5619-5674. Enzymatic Halogenation and Dehalogenation Reactions: Pervasive and Mechanistically Diverse. DOI:10.1021/acs.chemrev.6b00571. PMID:28106994.
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
inferred reaction step, hydride transfer, cofactor used
Step 2. Proton-coupled electron transfer (PCET) from FADH- to molecular oxygen and to the C4a-carbon atom of the isoalloxazine ring, leading to formation of FADH• and OOH• radicals. Inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
electron transfer, proton transfer, radical formation, cofactor used, intermediate formation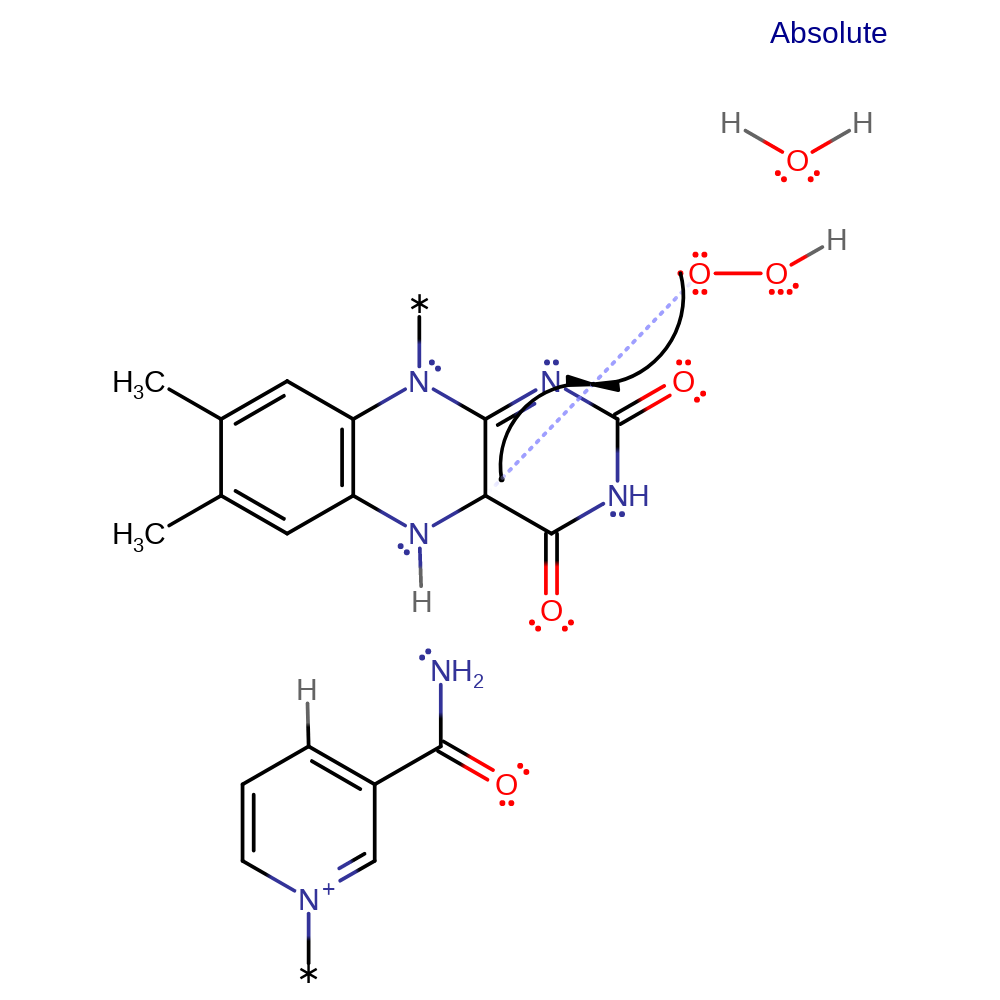
Step 3. FADH• and OOH• radicals colligate, in which each of the molecular fragments donate one of the bonding electrons, resulting in formation of C4a-hydroperoxyflavin (C4aOOH) intermediate. Inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
colligation, inferred reaction step, radical termination
Step 4. A chloride anion makes a nucleophilic attack on the flavin peroxide yielding hypochlorous acid (HClO) and C(4a) hydroxyflavin.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, intermediate formation, cofactor used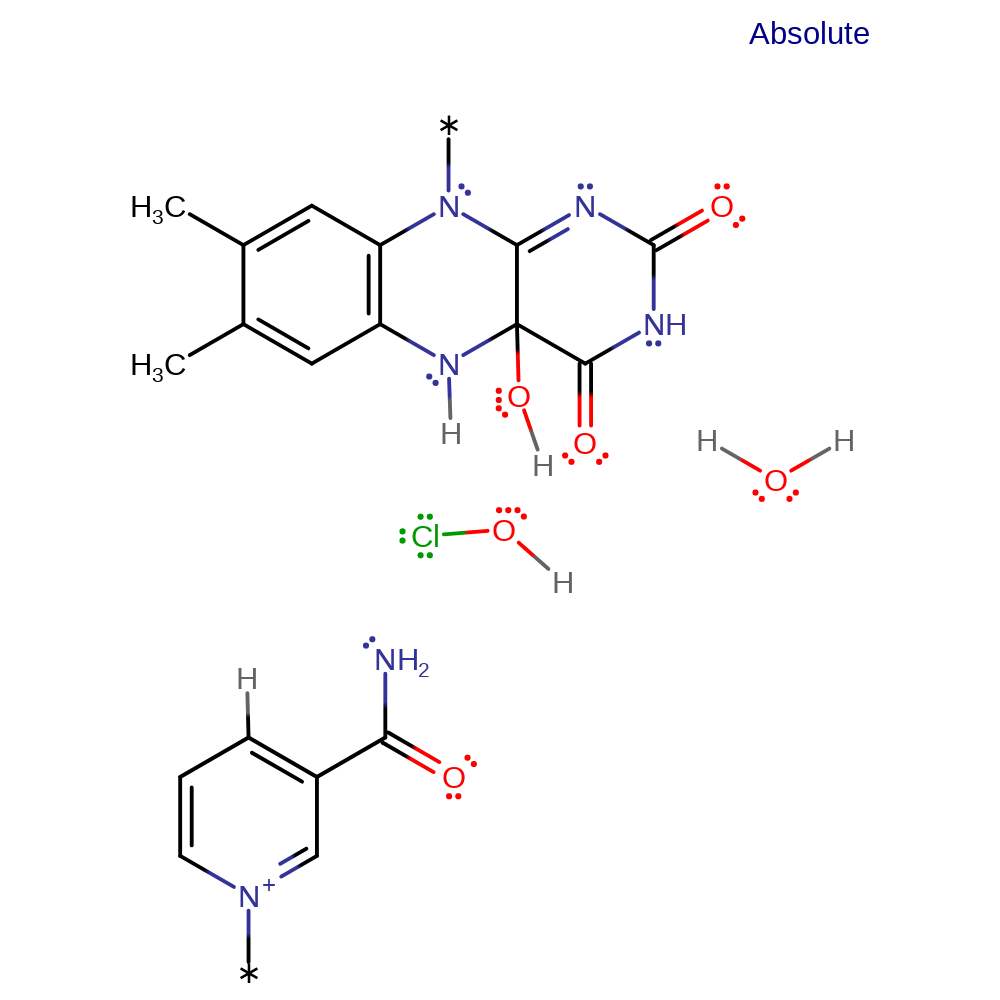
Step 5. Dehydration of C(4a) hydroxyflavin gives oxidised FAD (not shown). HOCl formed in the FAD binding site moves through a tunnel in the enzyme and becomes activated to chlorinate the substrate in the tryptophan binding site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
dehydration
Step 6. A hydrogen bonding interaction between HOCl and Lys79 activates HOCl by increasing its electrophilicity. The chlorine atom is inserted and forms a Wheland intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys79A | activator, hydrogen bond donor, proton donor |
Chemical Components
ingold: aromatic bimolecular electrophilic addition, proton transfer
Step 7. Deprotonation of the Wheland intermediate by Glu346 forms the product. This is rate-limiting.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu346A | proton acceptor |





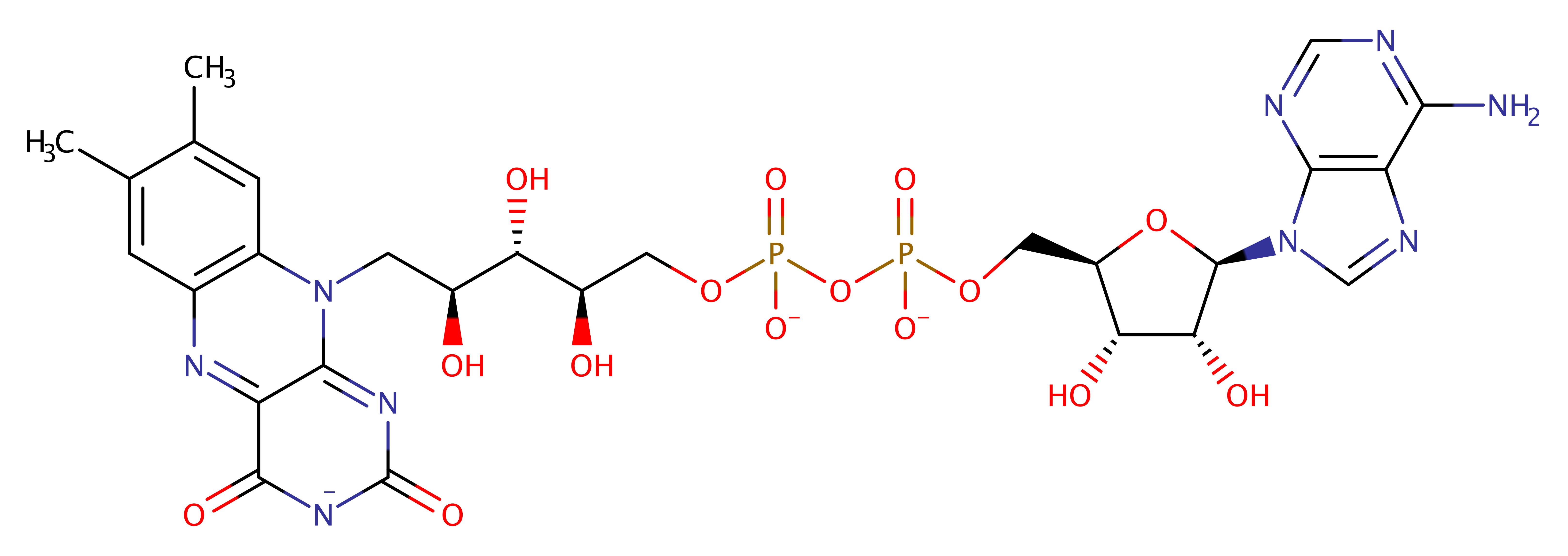


 Download:
Download: